Retracted: AT1R knockdown confers cardioprotection against sepsis-induced myocardial injury by inhibiting the MAPK signaling pathway in rats
Abstract
Myocardial dysfunction is an important manifestation of sepsis. In addition, inactivation of the mitogen-activated protein kinase (MAPK) signaling pathway has been reported to be beneficial in sepsis. The current study used gene expression profiling to demonstrate the overexpression of angiotensin II type 1 receptor (AT1R) and activation of the MAPK signaling pathway in sepsis. In this study, we used a rat model of sepsis established by cecal ligation and puncture to explore the mechanism of AT1R silencing in relation to the MAPK signaling pathway on myocardial injury. Various parameters including blood pressure, heart rate, and cardiac function changes were observed. Enzyme-linked immunosorbent assay was used to measure the concentration of cardiac troponin T (TnT), cardiac troponin I (cTnI), and creatine kinase isoenzyme muscle/brain (CK-MB). Myocardial enzyme, tissue antioxidant capacity, mitochondria swelling, and membrane potential were also detected. Terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling staining was applied to measure cell apoptosis, and messenger RNA and protein levels of apoptosis-related proteins (Fas ligand [Fasl], B-cell CLL/lymphoma [Bcl-2], p53) were also detected. Initially, sepsis rats exhibited decreased survival rate, but increased ejection fraction (EF), heart rate, and concentrations of TnT, cTnI, and CK-MB. Furthermore, decreased AT1R expression inactivated the MAPK signaling pathway (shown as decreased extracellular signal–regulated kinase and cyclic adenosine 3′,5′-monophosphate response element binding protein expression), decreased EF, heart rate, and concentrations of TnT, cTnI, and CK-MB, but increased sepsis rat survival rate. Eventually, decreased AT1R expression inhibited myocardial cell apoptosis (shown as decreased apoptosis rate and p53 and Fasl expression as well as increased Bcl-2 expression). These findings indicated that AT1R silencing plays an inhibitory role in sepsis-induced myocardial injury by inhibiting the MAPK signaling pathway.
1 INTRODUCTION
Sepsis is a host inflammatory response resulting from severe life-threatening infection with the presence of organ dysfunction.1 Myocardial depression is observed in approximately 80% of cases with severe sepsis, and 40% of myocardial depression can progress to myocardial injury, which ultimately affects the prognosis of patients.2 Myocardial injury in sepsis leads to the reduction of cardiac function, which is an essential mechanism underlying refractory hypotension and severely contributes to poor prognosis of sepsis.3 Nonspecific alterations in physiology are still used to define and diagnose sepsis, which include changes in respiratory rate, temperature, and heart rate.4 At present, sepsis treatments are mainly limited to drainage of the infected site or surgical removal, antimicrobial agents, and supportive care.5 Even though there have been considerable strides in antimicrobial development, sepsis is still linked to an unacceptably high mortality rate.6 The understanding of the etiology of sepsis must be of great therapeutic value and gene-related therapy might be a potential orientation.
Angiotensin II type 1 receptor (AT1R), a G-protein–coupled receptor, is known to mediate a majority of the recognized angiotensin II physiological actions, including renal tubular sodium transport, direct pressor response, and aldosterone secretion.7 AT1R is localized at chromosome 3q and is composed of five exons, of which the first four exons represent the 5′-untranslated region, whereas exon 5 is the only coding exon.8 Recently, it was found that AT1R is involved in cardiac hypertrophy development promoted by thyroid hormone.9 Further, inhibition of AT1R expression was reported to contribute to the protective roles of dietary polyphenols in cardiovascular conditions through the mitogen-activated protein kinase (MAPK)-dependent pathway.10 MAPKs, as the serine-threonine kinases, are involved in affecting cell growth, survival, proliferation, and differentiation by regulating relative genes.11, 12 A recent study revealed that heme oxygenase-1 could reduce the infiltration of neutrophils and prevent hepatic failure in sepsis through the inhibition of p38 MAPK signaling pathway.13 More importantly, inhibition of the MAPK signaling pathway could attenuate myocardial ischemic injury by functioning with kaempferol.14 Based on the above, it was hypothesized that AT1R could affect the MAPK signaling pathway, thereby influencing sepsis-induced myocardial injury. Therefore, the current study established a rat model of sepsis to elucidate the effects of AT1R gene silencing on myocardial injury of septic rats through the MAPK signaling pathway.
2 MATERIALS AND METHODS
2.1 Ethics statement
All animal experiments were approved by the animal ethics committee of Tianjin Huanhu Hospital. The use and the execution of experimental animals were strictly in accordance with the principles of the animal ethics committee, and all efforts were made to minimize the suffering of the included animals.
2.2 Model establishment and animal grouping
A total of 105 specific pathogen-free male rats (4-week old; calculated mean age 190 g ± 20 g) were purchased from Guangdong Medical Experimental Animal Center (Guangzhou, China). The rats were randomly assigned into the following seven groups (15 rats per group): the sham group (only laparotomy in rats), the model group (sepsis rats), the AT1R–small hairpin RNA (shRNA) group (sepsis rats transfected with AT1R-shRNA adenovirus expression vector), the negative control (NC) group (sepsis rats transfected with NC adenovirus expression vector), the Anisomycin group (sepsis rats treated with 5 mg/kg Anisomycin), the SB203580 group (sepsis rats treated with 5 mg/kg SB203580), and the AT1R-shRNA + Anisomycin group (sepsis rats transfected with AT1R-shRNA adenovirus expression vector and treated with 5 mg/kg Anisomycin). Anisomycin and SB203580 were purchased from Sigma-Aldrich Chemical Company (St Louis, MO). All rats were allowed free access to water and food and were raised under controlled conditions with a temperature of 20 ± 1°C, 60 ± 10% humidity, and lamplight from 6 am to 6 pm for a duration of 1 week.
All rats from each group were fasted for 12 hours before the operation. Next, the rats in each group were intraperitoneally anesthetized with 2% pentobarbital sodium (P3761; Sigma-Aldrich Chemical Company) using a dosage of 40 mg/kg. Sepsis rat models were established by cecal ligation and puncture.15 Rats in the model, AT1R-shRNA, and NC groups were anesthetized, given oxygen inhalation through a gas mask and fixed in the supine position with the limbs extended. Hair overlying the abdomen was shaved off using a razor blade, and iodophor disinfection was carried out for skin preparation. Under sterile conditions, the skin, muscle layers, and peritoneum were carefully incised (incision length about 1 cm) at a depth of about 0.3 cm below the midpoint of the white line of the abdomen. The muscle layers were clamped to directly expose the abdominal cavity. Next, the cecum was clipped using smooth forceps, and the mesocecum was carefully separated. The cecum was slowly taken out of the abdominal wall with smooth forceps, and the clipped cecum was pushed to the distal end. At approximately half distance from the ileocecal end, avoiding peripheral vessels, the cecum was ligated within the vascular arch of line 4 (to preserve smooth bowel movement). The cecum was punctured twice using a 10-mL syringe needle to squeeze the remained stool, and a 2-mm-wide and 1-mm-long strip rubber was set to prevent pinhole closure. Subsequently, the cecum was placed into the abdominal cavity, and the abdominal wall was sutured. Postanesthetization, the rats in the sham group only received laparotomy to move the cecum without ligation and puncture. Compound sodium chloride (50 mL/kg) was injected at the back of the neck immediately after operation. The rats were allowed access to water and forage 4 hours later. The success criteria for sepsis model establishment were as follows: the rats had decreased physical activity, decreased food intake, crouching lethargy, and canthus secretion. Seven hours after operation, positive blood culture indicated successful model establishment.16
2.3 Construction of shRNA-AT1R adenovirus expression vector
RNA interference technology was performed to construct shRNA adenovirus expression vector, which targeted AT1R. The small interfering RNA and its corresponding NC were designed on the basis of rat AT1R gene sequence, and then shRNA was synthetized (RiboBio Co, Ltd, Guangzhou, China). T-A cloning was used to connect the shRNA gene product of polymerase chain reaction (PCR) and T vector (pGEM-Teasy carrier system; Promega Company, Madison, WI). Blue-white screening was used to determine positive monoclonal. Next, the plasmids were extracted after amplification and sequenced after double-enzyme digestion. Restriction endonucleases, namely BglII (BGL-100; Cronin Biotechnology Co, Ltd, Beijing, China) and HindIII (MD202-02; Days Biochemical Technology Co, Ltd, Beijing, China) were used for double-enzyme digestion. Subsequently, the target gene fragment was cloned into pShuttle-CMV vector (AdEasy adenovirus vector system, 0119; Shanghai Ji Ran Biotechnology Co, Ltd, Shanghai, China) to construct plasmid pShuttle-CMV-rBD2. After linearizing, Escherichia coli BJ5183-AD-1 was transformed using electroporation, and recombinant adenovirus vector was constructed by homologous recombination with PAdEasy-1 carrying adenovirus genome.17
2.4 Transfection of adenovirus expression vector
Rats in each group were intraperitoneally anesthetized with 2% pentobarbital sodium (40 mg/kg). Next, the trachea cannula was dissected, and pure oxygen was administered to the rats using a DH-150 ventilator (Zhejiang Medical University Instrument Factory, Hangzhou, China). In the left fourth intercostal thoracotomy, a biepharostat was used for intercostal distraction to expose the surgical field. The pericardium was opened to expose the heart, aorta, and pulmonary artery root. Intact forceps were used to block circulation for 10 seconds at the distal end of the aorta and the pulmonary artery. The heart apex of rats in the AT1R-shRNA group was injected with 0.1 mL of Ad5-AT1R-shRNA and the NC group with 0.1 mL of Ad5-EGFP, whereas the sham group and the model group with 0.1 mL of diluted phosphate-buffered saline (PBS) containing recombinant virus. The changes in the heart were observed. During 10-s blocking circulation, the mice presented with gradually decreasing heart rate and decreased ventricular wall motion without ventricular fibrillation. However, the heart rate of rats gradually achieved normal levels after opening circulation. After transfection and withdrawal of the biepharostat, the left lung was dilated, and then the chest was closed. The muscle was injected with 100 thousand units of penicillin (CAS, 7177-48-2; Hangzhou Baisi Biotechnology Co, Ltd, Zhejiang, China). Postoperation, when the rats recovered spontaneous breathing levels, the ventilator and the unpin tracheal catheter were removed. After the operation, the rats were injected with 100 thousand units of penicillin daily for 3 days.18
2.5 Blood pressure, heart rate monitoring, and cardiac ultrasound detection
At the 1st, 3rd, and 5th day after transfection, rats in each group were intraperitoneally anesthetized with 2% pentobarbital sodium and fixed in the supine position. The right femoral artery was intubated, and an electrocardiograph monitor (LW9; Digicare Biomedical Technology, Inc, Florida) was used to detect blood pressure and heart rate in each group. In addition, HP Sonos-5500 echocardiography (probe frequency 2-4 MHz) (Philips, Bothell, WA) was adopted to detect the cardiac function, left ventricular ejection fraction (EF), and fractional shortening (FS) in each group. The heart rate fluctuation caused by anesthetics had little-to-no effect on the experiment, and brought about changes that were negligible in this study.19
2.6 Enzyme-linked immunosorbent assay
At the 1st, 3rd, and 5th day after transfection, venous blood samples (0.8-1.2 mL rat) were collected from all rats. The samples were centrifuged at 3000 rpm for 15 minutes at 4°C, and the supernatant was collected and stored at −80°C. The cardiac troponin T (TnT), cardiac troponin I (cTnI), and creatine kinase isoenzyme muscle/brain (CK-MB) in serum were detected according to specification provided by the R&D Company (Minneapolis, MN). Taking the TnT-α reaction as an example, the specific operations were as follows: the kit stored in the refrigerator at 4°C was taken out from the refrigerator and balanced for 15 to 30 minutes at room temperature. A 96-well plate was used to set up blank wells, standard wells, and sample wells. The blank well had the same steps as the other two wells without sample and enzyme reagent. Next, the standard well was added with 50 μL of standard samples of different concentrations. The sample well was added with 40 μL of sample diluent and 10 μL of test samples, avoiding touching the walls of the well after adding samples. The same steps were used for the blank well without the sample and enzyme reagent. After slight shaking for 1 minute, the samples were cultured avoiding light exposure at room temperature for 2 hours. The liquid inside the wells was discarded. The plate was rinsed with wash buffer (400 μL), and each well was added with 100 μL of rats TnT-α binding solution, and then incubated avoiding light exposure at room temperature for 2 hours. After rinsing with wash buffer (400 μL), each well was supplemented with 100 μL of substrate solution and incubated avoiding light exposure at room temperature for 30 minutes. Then, 100 μL of stop buffer was added to each well. A spectrophotometer (Beijing Instrument Factory, Beijing, China) was used to test the optical density (OD) of the samples at a wavelength of 450 nm. On the basis of standard and blank samples, the absorbance-concentration standard curve was plotted, and the concentration was calculated from its absorbance. Three duplicate wells were set up, and the experiment was repeated three times.20
2.7 Detection of serum myocardial enzymes
After 5 days of transfection, venous blood samples were collected from all rats. After the blood was centrifuged, the supernatant was discarded. The standard curve was plotted according to the instructions of the serum myocardial enzyme detection kit, and the activity of lactate dehydrogenase (LDH) and aspartate transaminase/glutamic-oxaloacetic transaminase (AST/GOT) were determined. The used kit was purchased from Nanjing Jianchen Bioengineering Co, Ltd (Nanjing, China).21
2.8 Detection of antioxidant capacity of myocardial tissues
Tissue homogenate was prepared using myocardial tissues of the left ventricle. The standard curve was plotted according to the instructions of the antioxidant capacity detection kit. Subsequently, the contents of malondialdehyde (MDA), total antioxidant capacity (T-AOC), and reduced glutathione (GSH) in myocardial tissues were measured. The used kit was purchased from Nanjing Jianchen Bioengineering Co, Ltd.22
2.9 Isolation, culture, and identification of rat myocardial tissues
A portion of the collected apical myocardial tissues was cleaned, sliced into 1 to 3 mm3 tissue blocks, treated with 6 g/L of protease at 37°C, centrifuged at 120 rpm for 30 minutes, and detached repeatedly until sufficient cells were obtained. Next, each detached cell suspension was transferred to Dulbecco modified Eagle medium (DMEM) medium containing horse serum to stop trypsin functioning. After centrifugation, a cell suspension was prepared using myocardial cell culture medium (500 mL of DMEM, 25 mL of fetal bovine serum, 50 mL of hydrogen sulfide, 6 mL of P/S) and incubated in a humidified incubator with 50 mL/L of CO2 at 37°C for 2 hours. After incubation, myocardial cells were separated using the differential adhesion method. Next, the cells were inoculated into a 6-well plate at a cell density of 3.5 × 106 cells/mL to further culture for 36 hours, followed by the first change of culture fluid. After 72 hours, the experiment was performed. At 72 hours before the culture, 0.1 mmol/L of 5- bromodeoxyuridine was added to the wells. Then, the primary myocardial cells were inoculated into a 6-well plate with sterile cover glass at a density of 105 cells/well to make the climbing glasses. After 3 days, the climbing glasses of cells were rinsed with PBS three times and fixed with 40 g/L of paraformaldehyde for 40 minutes, followed by three times PBS washing and sealing with 3 g/L of Triton X-100 at room temperature for 30 minutes. Next, the cells were rinsed with PBS three times and sealed with 200 μL of 50 g/L of bovine serum albumin for 1 hour at room temperature. The trinitrotoluene mouse monoclonal primary antibody (ab8295, 1 µg/mL; Abcam Inc, Cambridge, MA) was added onto the cover glass of myocardial cells for incubation at room temperature for 1.5 hours, followed by rinsing with PBS three times. The second antibody was added for incubation at room temperature for 2 hours avoiding light exposure, followed by rinsing with PBS three times. Finally, the cells were stained with 4′,6-diamidino-2-phenylindole, incubated at room temperature for 15 minutes and observed under a fluorescence microscope.23
2.10 Extraction of myocardial mitochondria and analysis of swelling and membrane potential in rats
After the rats were killed, about 100-mg myocardial tissue blocks were placed on ice, sliced into pieces, and added with a precooled homogenate medium A solution (including 250 mmol/L of sucrose, 0.1 mmol/L of ethylene glycol tetraacetic acid (EGTA), 2 mmol/L of 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), and 5 mg/L of calf serum protein, pH 7.2) to make 10% tissue homogenate on ice using a homogenizer using a 1:9 ratio. Next, the homogenate was centrifuged at 1500 rpm for 10 minutes at 4°C, and the precipitate was discarded. The supernatant was centrifuged at 10 000 rpm for 15 minutes at 4°C, and the precipitate was suspended using medium A solution. Then, the suspension was centrifuged at 10 000 rpm for 15 minutes at 4°C again. The precipitate was confirmed as myocardial mitochondria using transmission electron microscopy. The extracted mitochondria were suspended with B solution (200 mmol/L of sucrose, 0.01 mmol/L of EGTA, 10 mmol/L of HEPES, pH 7.4), and 0.5 mL of suspension was used for analysis of mitochondrial swelling via flow cytometry, with the ratio of average fluorescence intensity between the forward angle and the lateral angle (forward scatter/side scatter) indicating the swelling degree of the mitochondria. The mitochondrial suspension was quantified using the Coomassie brilliant blue method. And then, 1 mL above suspension was added with 1.5 μL of 1 mg/mL of prepared fluorescent dye JC-1 to conduct water bath at 37°C at constant temperature for 15 minutes avoiding light exposure, followed by centrifugation at 800g at normal temperature twice, in total 4 minutes. A flow cytometer was used to analyze the mitochondrial membrane potential of the supernatant. The mitochondrial membrane potential was expressed by the ratio of the average fluorescence intensity between the second channel and the first channel.23
2.11 Hematoxylin and eosin staining and Masson staining
Five days after injection, the rats were killed under anesthesia. A part of the apical myocardial tissues was fixed in 4% paraformaldehyde solution (AR1068; Boster Biological Technology Co, Ltd, Wuhan, China) for 48 hours. Then, the tissues were cleared in xylene after dehydration with graded alcohol from low to high concentrations (75%, 85%, and 95%). Next, the samples were embedded in melted paraffin. After cooling and solidification, the paraffin was sliced into four pieces (3-4 μm in thickness). Then, the samples were placed on glass slides after being ironed by hot water and dried at 45°C in an incubator. The sections were dewaxed with xylene twice, 20 minutes each time and rinsed with anhydrous alcohol twice, 1 minute each time. Next, graded alcohol from high to low concentrations (95%, 80%, and 70%) was used for dehydration, 1 minute each concentration, and distilled water was finally added to react for 1 minute. Two sections were stained using hematoxylin solution for 5 minutes and developed in acid water and ammonia for 10 seconds, followed by rinsing with water for 1 hour. After being dehydrated with alcohol from high to low concentrations (each for 10 minutes), the slices were stained with 5% ethanol eosin for 3 minutes, decolorized in 95% ethanol, cleared in xylene, and sealed with neutral balsam, followed by observation under an optical microscope (CX31; Olympus Optical Co, Ltd, Tokyo, Japan).20
Another two sections were stained with Masson. Next, the sections were stained with Weiger iron hematoxylin for 5 to 10 minutes and rinsed with water slightly. And then, the sections were differentiated with an ethanol solution and rinsed with water for several minutes, followed by staining with ponceau fuchsin acid liquid for 5 to 10 minutes and rinsed with distilled water slightly. After that, the sections were reacted with 1% phosphomolybdic acid for about 5 minutes until the collagen fiber or background was colorless, and the myelin sheath was observed to be red. The samples were stained with aniline blue solution for 5 minutes, dealt with 1% ice vinegar for 1 minute, and dehydrated using 95% ethanol several times. Following that, the samples were cleared with xylene after pure alcohol dehydration and sealed with neutral balata. The experiment was repeated three times. The results were observed and photographed under an optical microscope (CX31; Olympus Optical Co, Ltd).24
2.12 Immunohistochemistry
Five days after injection, the rats were killed under anesthesia. Next, apical myocardial tissues were rewarmed for 30 minutes at 37°C. The samples were sectioned in 3% H2O2 (80% carbinol) avoiding light exposure at room temperature for 10 minutes to eliminate endogenous peroxidase activity. Next, the sections were incubated in 10% goat serum at room temperature for 20 minutes. Then, the samples were incubated overnight with 50 μL of rabbit–anti-mouse AT1R rabbit monoclonal antibody (dilution ratio of 1:150, sc-579; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at 4°C and rewarmed for 45 minutes at 37°C. The samples were cultured with 40 to 50 μL of biotin-labeled goat–anti-rabbit immunoglobulin G second antibody (sc-2489; Santa Cruz Biotechnology, Inc) for 30 minutes at 37°C. After being rinsed with PBS three times (each time 5 minutes), the samples were added with 50 μL of horseradish peroxidase-labeled streptavidin (S-A/HRP), and developed with 3,3′-diaminobenzidine tetrahydrochloride (DAB) for 5 to 10 minutes, and the degree of dyeing was observed under a microscope. The samples were stained with hematoxylin for several seconds, differentiated with hydrochloric alcohol, and then rinsed under running water for 10 to 15 minutes. Next, 75%, 80%, 85%, 95%, and 100% gradient alcohol was used for sequential dehydration, xylene for verification, and neutral gum for sealing. The results were observed under an optical microscope. PBS solution instead of the primary antibody was set as NC, and positive histological section was considered as the positive control. Positive criteria were as follows: a total of five representative high-power fields (positive optical microscope; Nikon, Tokyo, Japan) were selected, with about 200 cells in each field, to count the number of positive cells with brown and yellow cell membranes. In each field, the gray value of AT1R positive cells = the number of AT1R positive cells ÷ the total cells. The gray value of AT1R positive cells in the whole slice was statistically analyzed using the mean ± standard deviation method.25
2.13 Terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling staining
Apical myocardial tissues were fixed in 4% neutral formaldehyde solution (10014128; Shanghai Rongbai Biological Technology Co, Ltd, Shanghai, China), dehydrated with ethanol gradually, followed by paraffin embedding, slicing, and dewaxing. A terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling (TUNEL) staining kit (TUN11684817; Beijing Jiamay Biotech Co, Ltd, Beijing, China) was used to test cell apoptosis. Then, the sections were treated for 10 minutes with 3% hydrogen peroxide methanol to remove catalase. Next, the sections were incubated with 20 μg/mL of proteinase K (MP Biomedical Corporation, CA) at room temperature for 15 minutes and rinsed with PBS twice (each time for 5 minutes). Simultaneously, the TUNEL reaction mixture was prepared. Then, the samples were incubated with 50 μL of TUNEL reaction mixture at 37°C for 1 hour and washed three times with PBS (each time for 5 minutes). Next, DAB (50 μL) was used for development, and the reaction was observed under a microscope. The reaction was stopped when the positive site was stained, and no obvious color was observed in the background. After staining, the samples were rinsed with distilled water several times. The apoptotic cells were cells stained brown in the nuclei. A total of 10 high-power fields of vision were randomly chosen from each group. The number of positive cells was counted in 1000 tumor cells to calculate the apoptosis rate as follows: the apoptosis rate (%) = (apoptotic cells/total cells) × 100%. The experiment was repeated three times.26
2.14 Reverse transcription quantitative PCR
Myocardial tissues (50 mg) preserved at −80°C were put into a 1.5-mL ribonuclease-free centrifuge tube. Total RNA content was extracted using a Trizol kit (Invitrogen Inc, Carlsbad, CA). Before reverse transcription, the precipitate was dissolved with DEPC, and the absorbance at 260 and 280 nm was determined. An OD260/OD280 value from 1.8 to 2.0 was considered as high-purity extraction. The RNA was reverse transcribed into complementary DNA (cDNA) in accordance with the instructions of the reverse transcription kit (6210A; TaKaRa Holdings Inc, Kyoto, Japan), with a total system of 10 μL. According to the fluorescent quantitation PCR kit (RR82LR; Takara Holdings Inc), the PCR reaction system was 20 μL, including 10 μL of SYBR Green, 0.8 μL of forward sequence (10 μM), 0.8 μL of reverse sequence (10 μM), 2 μL of cDNA (reverse transcription system), and 0.6 μL of distilled water. The messenger RNA (mRNA) levels of AT1R, angiotensin II (AngII), PI3K, Akt, Fas ligand (Fasl), B-cell CLL/lymphoma (Bcl-2), and p53 were detected with β-actin as the internal control. The relative mRNA levels were calculated using the method. The experiment was repeated three times. The quantitative PCR (qPCR) primer was synthetized by Beijing Genomics Institute (Shenzhen, China) (Table 1).20
| Gene | Primer sequence |
|---|---|
| AT1R | Forward: 5′-CTCTGACTAAATGGCTTACGACC-3′ |
| Reverse: 5′-TGACTACAGGGACAACAAAGGT-3′ | |
| AngII | Forward: 5′-TAGAAAGAGACCAGGGTGCCAAAGAGGG-3′ |
| Reverse: 5′-GCTTATCTCGCAGGGTCTTCTCATCCAC-3′ | |
| ERK | Forward: 5′-ATCAACACCACCTGCGACCTT-3′ |
| Reverse: 5′-TGGCCACATACTCGGTCAGAA-3′ | |
| CREB | Forward: 5′-CCACTCAGCCGGGTCTACCAT-3′ |
| Reverse: 5′-CCAGAGGCAGCTTGAACAACA-3′ | |
| p38 | Forward: 5′-CCAGGTACTGTGTCGGT-3′ |
| Reverse: 5′-CAGCGCATCGCCTTCTATCGCC-3′ | |
| Fasl | Forward: 5′-AAGAAGGACCACAACACAAATCTG-3′ |
| Reverse: 5′-CCCTGTTAAATGGGCCACACT-3′ | |
| Bcl-2 | Forward: 5′-CTACGAGTGGGATACTGGAGATGA-3′ |
| Reverse: 5′-ACAGCCAGGAGAAATCAAACAGA-3′ | |
| p53 | Forward: 5′-GCCGGCCTCTGACTTATTTCT-3′ |
| Reverse: 5′-CTCAGGTGGCTCATACGGTA-3′ | |
| β-Actin | Forward: 5′-CATCCTGCGTCTGGACCT-3′ |
| Reverse: 5′-TCAGGAGGAGCAATGATCTTG-3′ |
- Abbreviations: AngII, angiotensin II; AT1R, angiotensin II type 1 receptor; Bcl-2, B-cell CLL/lymphoma; CREB, cyclic adenosine 3′,5′-monophosphate response element binding protein; ERK, extracellular signal–regulated kinase; Fasl, Fas ligand.
2.15 Western blot analysis
Myocardial tissues (50 mg) were rinsed two times with PBS and supplemented with precooled RIPA lysis buffer (10 μL/mg). Next, the tissues were cut and placed in a homogenizer and then ground to a fine homogenate, followed by ultrasonic breaking. The whole aforementioned process was carried out on ice. Then, the samples were centrifuged at 12 000 rpm for 20 minutes at 4°C, and the supernatant was extracted and transferred to a 1.5-mL centrifuge tube. A BCA protein assay kit (Sigma-Aldrich Chemical Company) was used to detect protein concentration and adjust the protein concentration to ensure consistency in each group. Following adjustments, the protein extracts were quantified, added with 5 × SDS protein loading buffer (20 μL) and denatured at 95°C for 5 minutes. A total of 20 μL of samples (standard sample marker and denatured protein samples) were treated with SDS-PAGE electrophoresis; the process was carried out at a constant voltage of 80 V for 30 minutes until bromo blue dye reached the junction of two glues, and then 100-V voltage was used to continue electrophoresis for about 80 minutes. After electrophoresis, the glue was carefully removed and immersed in a transfer buffer, and the filter paper was also immersed into the transfer buffer. Next, the PVDF membrane was immersed in methanol for about 15 seconds in double-distilled H2O for 5 minutes and in the transfer buffer for 10 minutes. After taking out, the transfer device was installed from bottom to top as follows: filter paper, membrane, glue, and filter paper. During the operation, it was ensured that there were no bubbles in each layer. The protein was transferred to the membrane with a constant voltage of 15 V for 45 minutes. After that, the membrane was taken out and then blocked with 5% skimmed milk overnight at 4°C. After rinsing with Tris-buffered saline with Tween 20 (TBST), the samples were incubated overnight with primary monoclonal antibodies AT1R (ab124734, dilution ratio of 1:1000), AngII (ab108334, dilution ratio of 1:1000), total extracellular signal–regulated kinase (t-ERK)(1/2) (ab17942, dilution ratio of 1:1000), phospho-ERK (p-ERK)(1/2) (ab76299, dilution ratio of 1:8000), p38 (ab197348, dilution ratio of 1:1000), cyclic adenosine 3′,5′-monophosphate response element binding protein (CREB) (ab32515, dilution ratio of 1: 800), Fasl (ab68338, dilution ratio of 1:500), Bcl-2 (ab59348, dilution ratio of 1:500), and p53 (ab1101, dilution ratio of 1:500). All above-mentioned antibodies were purchased from Abcam Inc. After being rinsed with TBST, the samples were incubated with HRP second antibody (dilution ratio of 1:1000) for 1 hour at 37°C. After rinsing with TBST, enhanced chemiluminescence was applied for development of samples. The membranes were rinsed with running water and dried out in air. A charge-coupled device imaging system was applied for scanning, and the ImageJ software (NIH Image, Bethesda, MD) was used to analyze gray value, with β-actin as the internal control. The experiment was repeated three times.20
2.16 Statistical analysis
Statistical analyses were performed using the SPSS 21.0 software (IBM Corp, Armonk, NY). Measurement data were presented by mean ± standard deviation. First, the Kolmogorov-Smirnov method and the Levene method were used for test of normality and homogeneity test of variance. If data were in accordance with normal distribution and homogeneity test of variance, paired t test was used for analyzing comparisons within groups and unpaired t test was used for analyzing comparisons between groups. Differences between multiple groups were analyzed by single factor analysis of variance (ANOVA) or repeated measure ANOVA. In addition, the post hoc test was used for paired comparisons within groups. If data did not conform to normal distribution or homogeneity test of variance, the rank-sum test was conducted.
3 RESULTS
3.1 Rats exhibited worsening symptoms and increased death rate after modeling
Physiological conditions in each group were observed to ensure the success rate of sepsis rat modeling. After operation, the rats in the sham group presented with decreased physical activity shortly, but soon returned to be normal. The other physiological conditions were similar to those of preoperation. The rats in the model, NC, and AT1R-shRNA + Anisomycin groups presented with ingravescence, decreased activity, mental fatigue, glassy eyes, sleepiness, no thirst, poor appetite, low stimulus response, piloerection, ecphysesis, shiver, in addition to pyuria and diarrhea. These symptoms indicated toward successful establishment of sepsis models. The Anisomycin group exhibited more severe symptoms than the model and NC groups. The symptoms of rats in the AT1R-shRNA and SB203580 groups were similar to those of rats in the model and NC groups; however, the symptoms were relatively mild. The success rate of sepsis rat modeling was 100%. At 5 days after operation, there were no deaths in the sham group, seven rat deaths in the model group, eight rat deaths in the NC group, eight rat deaths in the AT1R-shRNA + Anisomycin group, two rat deaths in the AT1R-shRNA group, two rat deaths in the SB203580 group, and six rat deaths in the Anisomycin group (Table 2). The above results showed that worsening symptoms and elevated death rate were observed in sepsis rats.
| Item | Sham | Model | AT1R-shRNA | NC | Anisomycin | SB203580 | AT1R-shRNA + Anisomycin |
|---|---|---|---|---|---|---|---|
| Survival (n) | 15 | 8* | 13# | 7* | 9* | 13# | 7* |
| Death (n) | 0 | 7* | 2# | 8* | 6* | 2# | 8* |
- Abbreviations: AT1R, angiotensin II type 1 receptor; NC, negative control; shRNA, small hairpin RNA.
- n = 15; data were analyzed using χ2 test.
- * P < 0.05 vs the sham group.
- # P < 0.05 vs the model group.
3.2 AT1R silencing inhibits heart rate and increases EF value
To observe the changes in cardiac functions in rats from each group after modeling, echocardiography was used for relevant examinations. The results (Table 3) demonstrated that the EF value of the AT1R-shRNA and SB203580 groups was remarkably higher than that of the model, NC, and AT1R-shRNA + Anisomycin groups, but lower than that of the sham group (P < 0.05). No significant difference in the FS value was observed among all groups. An electrocardiogram monitor was used to observe the changes in blood pressure and heart rate in each group. Compared with the sham group, the heart rates of rats in the model, NC, and AT1R-shRNA + Anisomycin groups were found to be significantly increased, but decreased in the AT1R-shRNA and SB203580 groups. There were no significant differences in systolic and diastolic blood pressure among the four groups (P > 0.05). Therefore, heart rate was inhibited, and the EF value was promoted following AT1R silencing.
| Item | Sham | Model | AT1R-shRNA | NC | Anisomycin | SB203580 | AT1R-shRNA + Anisomycin |
|---|---|---|---|---|---|---|---|
| Heart rate, beats/min | 359.93 ± 23.32 | 458.00 ± 33.01* | 402.31 ± 23.38*# | 454.41 ± 35.05* | 521.33 ± 33.98*# | 404.54 ± 26.37*# | 461.29 ± 40.48* |
| Systolic pressure, kPa | 17.02 ± 1.62 | 16.35 ± 1.59 | 15.85 ± 1.38 | 15.87 ± 1.81 | 15.91 ± 1.70 | 15.90 ± 2.40 | 16.93 ± 1.48 |
| Diastolic pressure, kPa | 12.03 ± 1.31 | 11.86 ± 1.62 | 10.88 ± 1.45 | 11.13 ± 1.26 | 12.00 ± 1.18 | 12.27 ± 1.72 | 12.98 ± 1.56 |
| EF, % | 88.45 ± 10.71 | 55.07 ± 8.82* | 72.52 ± 7.68*# | 54.27 ± 8.01* | 40.12 ± 6.50*# | 70.90 ± 9.07*# | 54.42 ± 5.86* |
| FS, % | 49.68 ± 6.66 | 48.53 ± 6.07 | 48.07 ± 5.78 | 47.62 ± 6.04 | 44.05 ± 5.84 | 48.00 ± 4.05 | 44.82 ± 4.03 |
- Abbreviations: AT1R, angiotensin II type 1 receptor; EF, ejection fraction; FS, fractional shortening; NC, negative control; shRNA, small hairpin RNA.
- The sham group, only laparotomy in rats, n = 15; the model group, sepsis rats, n = 8; the AT1R-shRNA group, sepsis rats transfected with AT1R-shRNA adenovirus expression vector, n = 13; the NC group, sepsis rats transfected with NC adenovirus expression vector, n = 7; the Anisomycin group, sepsis rats treated with 5 mg/kg Anisomycin, n = 9; the SB203580 group, sepsis rats treated with 5 mg/kg SB203580, n = 13; the AT1R-shRNA + Anisomycin group, sepsis rats transfected with AT1R-shRNA adenovirus expression vector and treated with 5 mg/kg Anisomycin, n = 7; data of multiple groups were analyzed using one-way analysis of variance.
- * P < 0.05 vs the sham group.
- # P < 0.05 vs the model group.
3.3 Silencing AT1R inhibits TnT, cTnI, and CK-MB
Enzyme-linked immunosorbent assay was applied to detect biochemical markers related to cardiac function in rat serum to further reflect the rat cardiac function of each group. The TnT concentration in the model and NC groups was found to be significantly raised compared with that in the sham group at each time point (P < 0.05), whereas the TnT concentration decreased as time progressed in serum of rats with sepsis, and significant differences were detected between each time point (P < 0.05). The TnT concentration in the AT1R-shRNA and SB203580 groups was found to be significantly decreased, whereas the Anisomycin group presented with remarkably increased TnT concentration (P < 0.05). At 1 day after injection, cTnI concentration in the model and NC groups was found to be remarkably increased compared with that in the sham group (P < 0.05), and cTnI concentration increased significantly as time progressed, and significant differences were detected between each time point (P < 0.05). The AT1R-shRNA and SB203580 groups presented with significantly decreased cTnI concentration in serum of rats with sepsis (P < 0.05). Furthermore, elevated CK-MB concentration was detected in the model and NC groups (P < 0.05), and decreased significantly as time progressed. The AT1R-shRNA and SB203580 groups had markedly reduced cTnI and CK-MB concentration in serum of rats with sepsis (P < 0.05; Table 4). Therefore, it can infer that AT1R-shRNA could significantly decrease TnT, cTnI, and CK-MB concentration.
| Item | D 1 | D 3 | D 5 | |
|---|---|---|---|---|
| TnT, ng/mL | Sham group | 7.04 ± 1.27 | 6.98 ± 1.68 | 6.28 ± 1.08 |
| Model group | 14.84 ± 1.99* | 13.01 ± 1.02*& | 10.98 ± 1.01*&& | |
| AT1R-shRNA group | 11.06 ± 2.11*# | 9.41 ± 1.18*#& | 7.84 ± 1.07*#&& | |
| NC group | 15.01 ± 1.23* | 13.08 ± 1.13*& | 10.13 ± 1.01*&& | |
| Anisomycin group | 18.74 ± 1.51*# | 15.42 ± 1.09*#& | 13.52 ± 1.02*#&& | |
| SB203580 group | 11.07 ± 1.34*# | 9.31 ± 1.01*#& | 7.90 ± 1.04*#&& | |
| AT1R-shRNA + Anisomycin group | 15.34 ± 1.52* | 12.89 ± 1.42*& | 10.09 ± 1.17*&& | |
| cTnI, ng/mL | Sham group | 22.12 ± 2.01 | 23.14 ± 2.12 | 24.86 ± 2.07 |
| Model group | 101.12 ± 6.52* | 376.01 ± 28.64*& | 990.35 ± 52.38*&& | |
| AT1R-shRNA group | 55.41 ± 4.15*# | 76.41 ± 7.18*#& | 108.35 ± 9.48*#&& | |
| NC group | 113.54 ± 6.52* | 359.01 ± 24.17*& | 974.35 ± 25.41*&& | |
| Anisomycin group | 214.16 ± 8.41*# | 480.42 ± 37.47*#& | 1135.41 ± 45.52*#&& | |
| SB203580 group | 57.84 ± 4.82*# | 81.42 ± 7.46*#& | 120.13 ± 7.41*#&& | |
| AT1R-shRNA + Anisomycin group | 131.82 ± 6.94* | 364.15 ± 29.84*& | 964.17 ± 50.12*&& | |
| CK-MB, U/mL | Sham group | 7.54 ± 1.27 | 7.44 ± 1.54 | 7.39 ± 1.52 |
| Model group | 47.60 ± 8.99* | 34.62 ± 5.94*& | 23.26 ± 3.93*&& | |
| AT1R-shRNA group | 24.06 ± 3.18*# | 18.23 ± 1.98*#& | 13.06 ± 1.17*#&& | |
| NC group | 46.68 ± 7.59* | 33.61 ± 5.03*& | 21.78 ± 2.52*&& | |
| Anisomycin group | 70.15 ± 6.41*# | 56.41 ± 4.13*#& | 41.13 ± 3.08*#&& | |
| SB203580 group | 25.46 ± 2.13*# | 18.98 ± 1.52*#& | 13.56 ± 1.07*#&& | |
| AT1R-shRNA + Anisomycin group | 49.84 ± 4.12* | 32.71 ± 3.07*& | 20.13 ± 2.13*&& |
- Abbreviations: AT1R, angiotensin II type 1 receptor; CK-MB, creatine kinase isoenzyme muscle/brain; cTnI, cardiac troponin I; NC, negative control; P value is analysis of variance at different time points; TnT, troponin T.
- The sham group, only laparotomy in rats, n = 15; the model group, sepsis rats, n = 8; the AT1R-shRNA group, sepsis rats transfected with AT1R-shRNA adenovirus expression vector, n = 13; the NC group, sepsis rats transfected with negative control adenovirus expression vector, n = 7; the Anisomycin group, sepsis rats treated with 5 mg/kg Anisomycin, n = 9; the SB203580 group, sepsis rats treated with 5 mg/kg SB203580, n = 13; the AT1R-shRNA + Anisomycin group, sepsis rats transfected with AT1R-shRNA adenovirus expression vector and treated with 5 mg/kg Anisomycin, n = 7; the experiment was repeated three times and data of multiple groups were analyzed using one-way analysis of variance.
- * P < 0.05 vs the sham group.
- # P < 0.05 vs the model group.
- & P < 0.05 vs the first days after operation.
- & P < 0.05 vs the third day after operation.
3.4 Reduced serum LDH and AST after AT1R silencing
To further reflect the injury of myocardial function in each group, the serum myocardial enzyme detection kit was used to detect serum markers related to myocardial function injury. The results are shown in Table 5. Compared with the sham group, serum LDH and AST levels were found to be remarkably increased in the model, NC, AT1R-shRNA + Anisomycin, AT1R-shRNA, and SB203580 groups (P < 0.05). In comparison to the model and NC groups, the AT1R-shRNA and SB203580 groups presented with significantly reduced serum LDH and AST levels in rats with sepsis, whereas the Anisomycin group exhibited increased levels of serum LDH and AST (P < 0.05). The above results indicated that AT1R silencing decreased the levels of serum LDH and AST.
| LDH, U/L | AST, U/L | |
|---|---|---|
| Sham group | 251.15 ± 67.41 | 168.79 ± 30.13 |
| Model group | 642.17 ± 98.41* | 468.42 ± 52.46* |
| AT1R-shRNA group | 421.13 ± 71.12*# | 289.41 ± 46.84*# |
| NC group | 609.41 ± 98.42* | 502.17 ± 59.72* |
| Anisomycin group | 784.17 ± 100.01*# | 582.41 ± 89.74*# |
| SB203580 group | 408.45 ± 98.48*# | 301.52 ± 50.12*# |
| AT1R-shRNA + Anisomycin group | 658.41 ± 98.17* | 471.17 ± 52.11* |
- Abbreviations: AST, aspartate transaminase; AT1R, angiotensin II type 1 receptor; LDH, lactic dehydrogenase; NC, negative control; shRNA, short hairpin RNA.
- The sham group, only laparotomy in rats, n = 15; the model group, sepsis rats, n = 8; the AT1R-shRNA group, sepsis rats transfected with AT1R-shRNA adenovirus expression vector, n = 13; the NC group, sepsis rats transfected with negative control adenovirus expression vector, n = 7; the Anisomycin group, sepsis rats treated with 5 mg/kg Anisomycin, n = 9; the SB203580 group, sepsis rats treated with 5 mg/kg SB203580, n = 13; the AT1R-shRNA + Anisomycin group, sepsis rats transfected with AT1R-shRNA adenovirus expression vector and treated with 5 mg/kg Anisomycin, n = 7; the experiment was repeated three times and data of multiple groups were analyzed using one-way analysis of variance.
- * P < 0.05 vs the sham group.
- # P < 0.05 vs the model group.
3.5 Silencing AT1R reduces MDA while increasing GSH and T-AOC in sepsis rats
To further detect antioxidant capacity of rat myocardial tissues in each group, the antioxidant capacity detection kit was used to detect markers related to antioxidant capacity of myocardial tissues. The results are shown in Table 6. Compared with the sham group, significantly decreased GSH and T-AOC levels were observed in the model, NC, AT1R-shRNA + Anisomycin, AT1R-shRNA, and SB203580 groups (P < 0.05), in addition to significantly increased MDA levels. In comparison to the model and NC groups, the AT1R-shRNA and SB203580 groups presented with significantly reduced MDA levels while obviously elevated GSH and T-AOC levels in rats with sepsis, whereas the Anisomycin group demonstrated opposite trends with the AT1R-shRNA and SB203580 groups (P < 0.05). The above results showed that AT1R silencing resulted in decreased MDA levels, in addition to elevated GSH and T-AOC levels.
| MDA, nmol/mg | GSH, kU/g | T-AOC, kU/g | |
|---|---|---|---|
| Sham group | 3.18 ± 0.41 | 23.65 ± 2.93 | 4.28 ± 0.34 |
| Model group | 7.97 ± 0.53* | 14.81 ± 1.45* | 2.09 ± 0.21* |
| AT1R-shRNA group | 5.38 ± 0.70*# | 18.41 ± 2.13*# | 3.15 ± 0.31*# |
| NC group | 8.01 ± 0.56* | 14.89 ± 2.09* | 2.13 ± 0.22* |
| Anisomycin group | 12.48 ± 2.01*# | 10.46 ± 2.17*# | 1.58 ± 0.16*# |
| SB203580 group | 5.07 ± 0.64*# | 18.04 ± 2.34*# | 3.75 ± 0.32*# |
| AT1R-shRNA + Anisomycin group | 8.12 ± 0.64* | 15.01 ± 2.44* | 2.15 ± 0.28* |
- Abbreviations: AT1R, angiotensin II type 1 receptor; GSH, glutathione; MDA, malondialdehyde; NC, negative control; T-AOC, total antioxidant capacity; shRNA, short hairpin RNA.
- The sham group, only laparotomy in rats, n = 15; the model group, sepsis rats, n = 8; the AT1R-shRNA group, sepsis rats transfected with AT1R-shRNA adenovirus expression vector, n = 13; the NC group, sepsis rats transfected with negative control adenovirus expression vector, n = 7; the Anisomycin group, sepsis rats treated with 5 mg/kg Anisomycin, n = 9; the SB203580 group, sepsis rats treated with 5 mg/kg SB203580, n = 13; the AT1R-shRNA + Anisomycin group, sepsis rats transfected with AT1R-shRNA adenovirus expression vector and treated with 5 mg/kg Anisomycin, n = 7; the experiment was repeated three times and data of multiple groups were analyzed using one-way analysis of variance.
- * P < 0.05 vs the sham group.
- # P < 0.05 vs the model group.
3.6 Identification of myocardial cells
Immunofluorescence staining was used to detect cTnT expression in myocardial cells. After culture for 5 days, primarily cultured rat myocardial cells had been made into tissue blocks. Myocardial cell specific marker protein cTnT, which was labeled by immunofluorescence, was observed as green fluorescence in the cytoplasm, indicating its positive expression. No expression of cTnT was observed in nonmyocardial cells. The purity of myocardial cells was calculated to be 95 ± 0.62%, which was high, as shown in Figure 1.
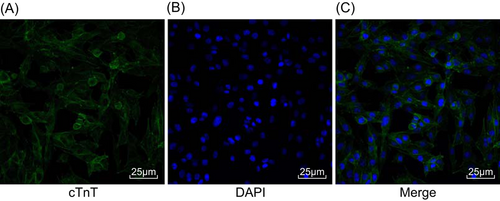
Positive expression of cTnT was observed in myocardial cells. A, Green FITC–labeled cTnT; B, blue DAPI–labeled nucleus; C, the combination of A and B was shown. FITC, fluorescein isothiocyanate; DAPI, 4′,6-diamidino-2-phenylindole
3.7 Slightly swollen mitochondria of myocardial cell and no vacuoles are found after silencing AT1R
An electron microscope was used to observe mitochondrial morphology. It was found that the sham group presented with normally shaped mitochondria, with clear boundaries, homogeneous matrix, and dense mitochondria cristae. The model, NC, and AT1R-shRNA + Anisomycin groups showed swollen mitochondria, decreased matrix density, broken cristae, and concentric circles. Mitochondria in the AT1R-shRNA and SB203580 groups were observed to be slightly swollen, with no vacuoles or a little vacuolization, and no significant decrease in the number of mitochondria. The Anisomycin group presented with swollen mitochondria, decreased matrix density, and vacuolization of mitochondria (Figure 2).
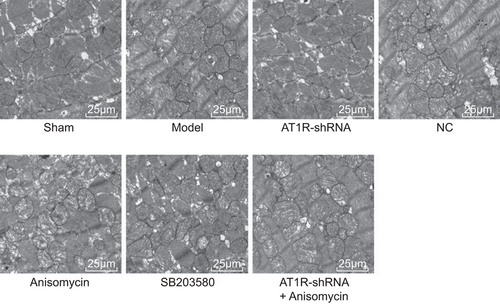
Silencing AT1R reduced swelling and vacuole of mitochondria. AT1R, angiotensin II type 1 receptor; NC, negative control; shRNA, short hairpin RNA
3.8 Silencing AT1R can reduce the degree of mitochondrial and increase membrane potential
To further reflect mitochondria swelling and membrane potential of myocardial cells in each group, related detection was performed using flow cytometry. Compared with the sham group, the model, NC, AT1R-shRNA + Anisomycin, AT1R-shRNA, and SB203580 groups presented with obviously increased mitochondrial swelling, in addition to significantly decreased membrane potential (P < 0.05). In comparison to the model and NC groups, the AT1R-shRNA and SB203580 groups had significantly reduced mitochondrial swelling while obviously elevated membrane potential was observed in rats with sepsis, and the Anisomycin group presented with opposite trends with the AT1R-shRNA and SB203580 groups (P < 0.05; Table 7). The above results showed that mitochondria swelling could be reduced and membrane potential could be elevated by silencing AT1R.
| FSC/SSC (swelling) | FL2/FL1 (membrane potential) | |
|---|---|---|
| Sham group | 0.50 ± 0.06 | 1.98 ± 0.21 |
| Model group | 0.75 ± 0.07* | 1.42 ± 0.18* |
| AT1R-shRNA group | 0.60 ± 0.07*# | 1.75 ± 0.24*# |
| NC group | 0.81 ± 0.07* | 1.64 ± 0.13* |
| Anisomycin group | 0.98 ± 0.10*# | 1.15 ± 0.12*# |
| SB203580 group | 0.59 ± 0.07*# | 1.74 ± 0.15*# |
| AT1R-shRNA + Anisomycin group | 0.84 ± 0.08* | 1.50 ± 0.13* |
- Abbreviations: AT1R, angiotensin II type 1 receptor; FL2/FL1, fluorescence intensity between the second channel and the first channel; FSC, forward scatter; NC, negative control; shRNA, short hairpin RNA; SSC, side scatter.
- n = 7; the experiment was repeated three times and data of multiple groups were analyzed using one-way analysis of variance.
- * P < 0.05 vs the sham group.
- # P < 0.05 vs the model group.
3.9 Myocardial histopathological changes are mitigated by AT1R silencing
Hematoxylin and eosin staining was performed to observe the pathological structure of rat myocardial tissues, and the results are shown in Figure 3A. The staining results showed that cardiac muscle fibers were complete, and cells were neatly arranged in the sham group. Furthermore, the fibers were interconnected reticulate, and the nucleus was clear and uniform in size. In addition, the cytoplasm was uniformly stained, and caryotheca was complete and there was no cellular swelling. In the model, NC, and AT1R-shRNA + Anisomycin groups, myocardial fibers were found to be swollen, irregularly arranged, irregularly shaped, with dissolution, small stain in cytoplasm, and accompanied by inflammatory cell infiltration. The Anisomycin group showed relatively more swollen myocardial fibers, more irregular shapes, more disordered arrangement, dissolution and lightly stained cytoplasm, accompanied by a large number of inflammatory cells. In the AT1R-shRNA and SB203580 groups, cardiac muscle fibers had a certain swelling, deeply stained nuclear, low alignment, minor dissolution, and slightly stained cytoplasm with inflammatory cell infiltration.
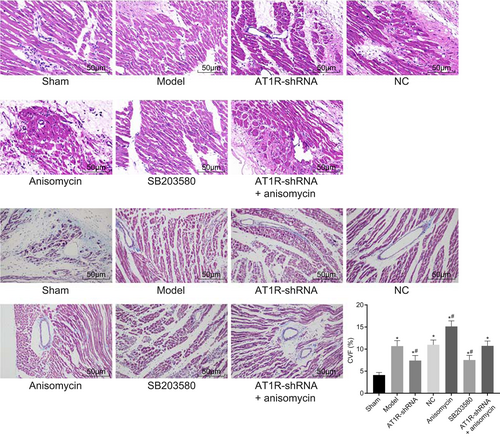
AT1R silencing reduced myocardial histopathological changes. A, HE staining was used to observe pathological structure of myocardial tissues (×200). B, Masson staining was used to observe collagen fibrils in myocardial tissues (×200); the experiments were repeated three times and data of multiple groups were analyzed using one-way analysis of variance. *P < 0.05 vs the sham group; #P < 0.05 vs the model group. AT1R, angiotensin II type 1 receptor; HE, hematoxylin and eosin; NC, negative control; shRNA, short hairpin RNA
Masson staining was conducted to observe the arrangement and proliferation of collagen fibers in rat myocardial tissues in each group. The results (Figure 3B) revealed that cardiac muscle cells were arranged orderly, and only a small amount of blue collagen fibers were visible in myocardial gap in the sham group. In the model, NC, and AT1R-shRNA + Anisomycin groups, a large number of blue collagen fibers were observed in the lesions. Collagen precipitation was found to be remarkable increased in the AT1R-shRNA and SB203580 groups, and furthermore, the proliferation of collagen fibers in the interstitium was reduced, and the collagen fibers were arranged disorderly. The Anisomycin group presented with significantly increased collagen deposition, obviously proliferated collagen fibers in the interstitium and more disorderly arranged collagen fibers. Therefore, these results revealed that AT1R silencing alleviated myocardial histopathological changes.
3.10 Increased AT1R protein levels contribute to the occurrence of sepsis
AT1R levels were measured in each group by immunohistochemistry to investigate the role played by AT1R in the occurrence of sepsis. AT1R protein levels were localized on the cell membrane, and positive staining was observed as brown precipitation on the cell membrane. Immunohistochemical results demonstrated that rat endothelial cells and myocardium fibroblasts presented with a small amount of AT1R positive expression in the sham group. Compared with the sham group, in the model group, cardiac muscles revealed more scattered necrosis and were confined to the endocardium, and the positive expression of endothelial cells and myocardium fibroblasts were found to be increased (P < 0.05). In the AT1R-shRNA and AT1R-shRNA + Anisomycin groups, AT1R expression in the cardiac muscle showed a decreasing trend, and the AT1R positive staining area and staining power both declined (Figure 4A). Compared with the sham group, AT1R protein levels in the model, NC, Anisomycin, and SB203580 groups were found to be significantly increased, but decreased protein levels were observed in the AT1R-shRNA and AT1R-shRNA + Anisomycin groups (all P < 0.05). Compared with the model group, AT1R protein levels in the AT1R-shRNA and AT1R-shRNA + Anisomycin groups remarkably declined (P < 0.05). No significant protein level differences were found in the model, NC, Anisomycin, and SB203580 groups (P > 0.05; Figure 4B). Therefore, sepsis was accompanied by increasing AT1R protein levels.
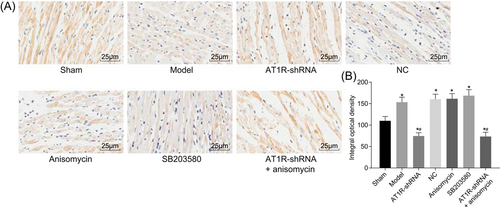
Increased AT1R protein levels in sepsis. A, Immunohistochemistry was used to detect AT1R (×400). B, Histogram of AT1R positive expression in myocardial tissue of rats in each group; the experiment was repeated three times and data of multiple groups were analyzed using one-way analysis of variance. *P < 0.05 vs the sham group; #P < 0.05 vs the model group. AT1R, angiotensin II type 1 receptor; NC, negative control; shRNA, short hairpin RNA
3.11 AT1R silencing inhibits myocardial cell apoptosis
TUNEL staining was used to detect myocardial cell apoptosis in each group. The results were observed using a light microscope, and myocardial nuclei in the sham group were observed to be stained blue, with clear and complete boundaries. A small number of apoptotic nuclei were seen in brown color or as brown particles. Compared with the sham group, myocardial cell apoptosis in other groups was found to be significantly increased (all P < 0.05). In comparison to the model, NC, and AT1R-shRNA + Anisomycin groups, the Anisomycin group presented with increased myocardial cell apoptosis, whereas the AT1R-shRNA and SB203580 groups had decreased myocardial cell apoptosis (all P < 0.05). There were no significant differences between the sham and AT1R-shRNA groups (Figure 5). Moreover, myocardial cell apoptosis in the model and NC groups showed no significant differences (P > 0.05). The above findings showed that myocardial cell apoptosis was suppressed after AT1R silencing.
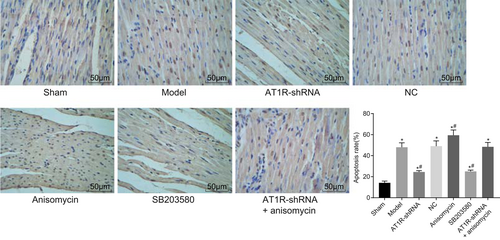
AT1R gene silencing reduced myocardial cell apoptosis after myocardial injury induced by sepsis (×200). The experiment was repeated three times and data of multiple groups were analyzed using one-way analysis of variance. *P < 0.05 vs the sham group; #P < 0.05 vs the model group. AT1R, angiotensin II type 1 receptor; NC, negative control; shRNA, short hairpin RNA
3.12 AT1R silencing inhibits the MAPK signaling pathway and apoptosis
Lastly, reverse transcription qPCR (RT-qPCR) and Western blot analyses were used to detect the levels of AT1R, AngII, the MAPK signaling pathway and apoptosis-related genes. As shown in Figure 6, compared with the sham group, the AT1R Mrna and protein levels were found to be notably raised in the other groups, whereas AngII mRNA and protein levels in each group showed no significant changes (all P > 0.05). Compared with the sham group, the mRNA levels of p53, Fasl, AT1R, ERK, p38, and CREB were found to be increased, whereas the mRNA level of Bcl-2 decreased in the other groups (all P < 0.05). The model, NC, Anisomycin, and SB203580 groups exhibited significantly increased AT1R mRNA and protein levels, whereas the AT1R-shRNA and AT1R-shRNA + Anisomycin groups had remarkably decreased AT1R mRNA and protein levels. In comparison to the model group, the AT1R-shRNA and AT1R-shRNA + Anisomycin groups presented with obviously reduced AT1R levels.
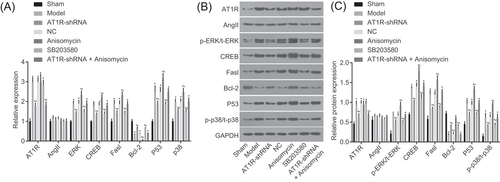
RT-qPCR and Western blot analysis indicated that silencing AT1R suppressed the MAPK signaling pathway and apoptosis in rat myocardial tissues. A, mRNA expression of AT1R, AngII, MAPK signaling pathway-related factors, and apoptosis-related factors detected by RT-qPCR. B, Protein expression of AT1R, AngII, MAPK signaling pathway-related factors, and apoptosis-related factors detected by Western blot analysis. C, Gray values of AT1R, AngII, p-ERK/t-ERK, CREB, Fasl, Bcl-2, and p53 were detected by Western blot analysis. The sham group, only laparotomy in rats, n = 15; the model group, sepsis rats, n = 8; the AT1R-shRNA group, sepsis rats transfected with AT1R-shRNA adenovirus expression vector, n = 13; the NC group, sepsis rats transfected with negative control adenovirus expression vector, n = 7; the Anisomycin group, sepsis rats treated with 5 mg/kg Anisomycin, n = 6; the SB203580 group, sepsis rats treated with 5 mg/kg SB203580, n = 12; the AT1R-shRNA + Anisomycin group, sepsis rats transfected with AT1R-shRNA adenovirus expression vector and treated with 5 mg/kg Anisomycin, n = 7; the experiment was repeated three times and data of multiple groups were analyzed using one-way analysis of variance. *P < 0.05 vs the sham group; #P < 0.05 vs the model group. AngII, angiotensin II; AT1R, angiotensin II type 1 receptor; Bcl-2, B-cell CLL/lymphoma; CREB, cyclic adenosine 3′,5′-monophosphate response element binding protein; ERK, extracellular signal–regulated kinase; Fasl, Fas ligand; MAPK, mitogen-activated protein kinase; mRNA, messenger RNA; NC, negative control; RT-qPCR, reverse transcription quantitative polymerase chain reaction; shRNA, short hairpin RNA
Compared with the model, NC, and AT1R-shRNA + Anisomycin groups, the mRNA levels of p53, Fasl, p38, ERK, and CREB were found to be significantly decreased, whereas Bcl-2 mRNA levels were significantly increased in the AT1R-shRNA and SB203580 groups (all P < 0.05). The Anisomycin group presented with opposite trends of these mRNA levels with the AT1R-shRNA and SB203580 groups. There were no significant differences in the model, NC, and AT1R-shRNA + Anisomycin groups (all P > 0.05). Compared with the sham group, the protein levels of p53, Fasl, AT1R, p-ERK/t-ERK, p-p38, and CREB were found to be significantly increased, whereas Bcl-2 protein levels were obviously decreased in the other groups (all P < 0.05). Compared with the model, NC, and AT1R-shRNA + Anisomycin groups, the protein levels of p53, Fasl, AT1R, p-ERK/t-ERK, p-p38, and CREB were found to be significantly decreased, whereas Bcl-2 protein levels were significantly increased in the AT1R-shRNA and SB203580 groups (all P < 0.05). The Anisomycin group exhibited opposite trends of these protein levels with the AT1R-shRNA and SB203580 groups. There were no significant differences in the model, NC, and AT1R-shRNA + Anisomycin groups (all P > 0.05). These data indicated that AT1R silencing could suppress the MAPK signaling pathway and myocardial cell apoptosis.
4 DISCUSSION
Sepsis is a highly lethal disease, characterized by extensive apoptosis-induced immune cell depletion and profound immunosuppression development state.27 Symptomatically, sepsis is often clinically manifested by two or more of the following symptoms: fever or hypothermia, tachypnea, leukocytosis or leukopenia, and tachycardia; furthermore, sepsis leads to one or more severe complications, including coma, renal failure, intravascular coagulation, hypotension, and cardiac failure.28 Interestingly, a previous study found that deregulated apoptosis and overshooting neutrophil functions can impair immune and organ functions in both sepsis and multiple organ failure.29 Aiming to explore the roles of AT1R gene silencing in myocardial injury of septic rats through the MAPK signaling pathway, the study conducted a series of experiments, and finally concluded that AT1R gene silencing protected septic rats against myocardial injury by inhibiting the activation of the MAPK signaling pathway.
We obtained the first conclusion that AT1R silencing mediated by shRNA attenuated cardiac dysfunction and was protective against sepsis-induced myocardial injury. In addition, the current study noted an increase in cTnI levels, indicating that there was myocardial damage when sepsis occurred. cTnI is a known indicator that reflects the degree of myocardial injury after sepsis and can be further used to measure protective effects of drug intervention with high sensitivity and specificity.30, 31 In accordance with the current study, Fuchs et al32 reported that AT1R was overexpressed in cases of postmyocardial infarction. Furthermore, it is well documented that AT1R is a key compound of a renin-angiotensin-aldosterone system (RAAS), and declined RAAS was previously demonstrated to reduce endothelial injury and inflammatory response in sepsis.33, 34 In addition, another study revealed that inhibition of AT1R could suppress the inflammatory response induced by Aah venom, in the heart and the aorta by declining the metabolic enzymes (CK and CK-MB) in sera.35 In addition, Yue et al36 found that chronic activation of AT1R decreased STAT3 expressions, resulting in nuclear accumulation of U-STAT3, possible contributing to the progression of cardiac hypertrophy that increases myocardial cell susceptibility to ischemia/reperfusion (I/R) injury. Similarly, inhibition of AT1R was demonstrated to reduce myocardial injury during reperfused myocardial infarction.37 Moreover, a recent study demonstrated that the role played by AT1R in cardiac dysfunction and suppression of AT1R expression was beneficial in preventing myocardial injury during sepsis. In addition, Cui et al38 reported the inhibition of AT1R-improved cardiac function in rats with chronic heart failure. Collectively, the findings of the current study indicated that AT1R might be an important target for the treatment of sepsis, but further researches on its underlying mechanism are required in future.
Interestingly, the current study subsequently discovered that the protective role of AT1R gene silencing in sepsis-induced cardiac dysfunction and myocardial injury may be achieved through inactivation of the MAPK signaling pathway. Furthermore, the current study reported that AT1R gene silencing inhibits the activation of the MAPK signaling pathway in sepsis. In line with our finding, previously, AT1R was demonstrated to induce extracellular signal–regulated kinases (ERK1/2) and p38 MAPK.33 In addition, it has been reported that inhibition of the AT1R-ERK/p38 MAPK signaling pathway relieves diabetes-induced cardiac dysfunction and improves inflammatory response.39 Furthermore, activation of the AT1R/p38 MAPK signaling pathway was associated with myocardial injury in myocardial infarction-induced rat models, indicating that AT1R silencing and inactivation of the MAPK signaling pathway may contribute to reducing myocardial injury.40 Additionally, the current study also found that inhibition of the MAPK signaling pathway suppresses inflammatory response and myocardial injury in sepsis. Similarly, a recent study noted that inhibition of the p38-MAPK signaling pathway by oxymatrine could reduce sepsis-induced myocardial injury.3 Another study noted that declined proinflammatory cytokines and inactivation of the p38 MAPK signaling pathway caused by irbesartan could contribute to attenuated cardiac dysfunction during sepsis.41 Taken together, we conjectured that AT1R gene silencing inactivated the MAPK signaling pathway, thus leading to improved cardiac function and a reduction in myocardial cells in sepsis.
In conclusion, the current study suggests that AT1R gene silencing protects septic rats from myocardial injury by suppressing the activation of the MAPK signaling pathway. MAPK inactivation increases antioxidant activity, alleviates inflammation, and inhibits myocardial cell apoptosis, contributing to the attenuation of myocardial depression in sepsis. Our study warrants an in-depth examination of the clinical use targeting AT1R in the treatment of sepsis-induced cardiac dysfunction and myocardial injury.
ACKNOWLEDGMENTS
We would like to thank sincerely the reviewers for critical comments on this study.
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.