Integrating genomics and conservation to safeguard plant diversity
整合基因组学与保护生物学研究,保护植物多样性
Abstract
enConserving genetic diversity has been increasingly recognized as a keystone towards the protection of biodiversity and natural resources for future generations of humankind. In turn, the ongoing genomic revolution provides us with the technical innovations and expanded knowledge required to establish programs to integrate genomic approaches into programs to protect plant diversity and its genetic diversity. Although obtaining whole genomes of all organisms is still a far-fetched dream, rapid progress in assembling whole genomes across the plant tree-of-life has provided access to reference genomes for many plants but, unfortunately, not all lineages of the plant tree-of-life. Addressing the existing taxon gaps is arguably best achieved by focusing on the genomes of plant species that experience high extinction threats or play a crucial role in ecosystem restoration. This review summarizes not only the recent progress in plant genomics with an emphasis on their importance to the conservation of plant diversity but also outlines steps to integrate genomics into plant conservation. Given the complexity of threats driving plant extinction, such as reduced genetic diversity, enhanced genome erosion, and loss of species integrity by hybridization, the integration of genomic research into our conservation efforts is crucial to understand extinction processes and, most importantly, to support the targets set to achieve “Harmony with Nature.”
摘要
zh遗传多样性保护越来越被认为是为人类后代保护生物多样性和自然资源的基石。而且,正在进行的基因组革命为我们提供了技术创新,以及实施发现与保存遗传多样性项目所需的基本信息。尽管获得所有生物的全基因组仍是一个梦想,但植物生命之树全基因组的快速组装为许多植物谱系提供了大量参考基因组。通过研究受威胁植物和对生态系统恢复至关重要的植物的基因组,解决现有的分类问题,将为我们实现“人与自然和谐共生”的愿景提供重要信息。本文综述了植物基因组学的研究进展,强调了其对植物多样性保护的重要意义,并建议构建对生物多样性保护具有较高价值的植物的基因组。
Practitioner points
en
-
Successful protecting of biodiversity requires the conservation of species diversity and must be based on protection of genetic diversity.
-
The genomic revolution provides access to innovations that do not only transform agroforestry practices but will also revitalize our efforts to protect plant diversity, including the natural resources provided by plants.
-
The review agures for two initiatives focusing on the genomes of either threatened species (= Threatened Species Initiative) or species playing a key role in ecosystem restoration.
实践者要点
zh
-
成功的生物多样性保护不仅需要保护物种多样性,还必须以保护遗传多样性为基础。
-
基因组革命带来的技术创新不仅变革了农林业的实践,还将为我们保护植物资源及其多样性注入新的活力。
-
本综述重点关注受威胁物种和生态系统恢复关键物种的基因组。
1 INTRODUCTION
By considering current progress in plant genomics, especially the rapid expansion of the number of plant genomes available, this study takes forward arguments to integrate genomics into our efforts to protect plant diversity and the natural resources provided as a cornerstone to achieving “Harmony with Nature.” The review was written in the context of the enhanced recognition of the urgent need to protect genetic diversity. In turn, the rapidly increased number of reference plant genomes provides a remarkable resource to support the conservation of threatened species. Fully sequenced and annotated genomes have been made available for only a small number of threatened species or their close relatives. Thus, efforts are needed not only to utilize the already available reference genomes but also to expand the coverage of the reference genomes across all branches of the tree of life. To enhance the impact of genomics in our efforts to preserve plant diversity and restore ecosystems, special focus is required to be given to threatened and keystone species of major habitats considered to be under threat, such as the rainforest of Southeast Asia, besides taxa considered to be the key in the efforts to establish healthy ecosystems. In general, the review aims to highlight the urgent need to focus on research questions concerning the genomics of extinction and preservation of genomic diversity of threatened species, to support not only the conservation of single species but also to sustain our efforts of ecosystem restoration.
2 RATIONALE OF THE ARGUMENT
With the rapid increase of anthropogenic threats to ecosystems, innovative and visionary approaches are urgently needed to reduce the negative trade-offs to achieve the UN target of “Harmony with Nature” (https://sdgs.un.org/2030agenda; https://www.unep.org/news-and-stories/story/towards-vision-2050-biodiversity-living-harmony-nature). Protecting biodiversity has been widely recognized to be among the most critical cornerstones to establish an ecological civilization but in turn halting the ongoing biodiversity loss is among the biggest challenges faced by humanity (https://news.un.org/en/story/2021/10/1102942). Among the major advancements in biological sciences, genomics stands out (Hogg et al., 2022; Kress et al., 2022). Insights provided by the access to the genetic blueprint via whole genome sequencing have been considered a game changer in conservation biology and restoration ecology (e.g., Breed et al., 2019; Formenti et al., 2022; Hogg et al., 2022; Mohr et al., 2022; O'Brien 2022; Shafer et al., 2015; Supple & Shapiro, 2018). The current contribution of genomics to species protection range from documenting the loss of genetic diversity in domestic and wild species, integrating genetic diversity to protect species via breeding programs, and even rescue genetics to secure the genomic health of protected species (Ralls et al., 2020). Unfortunately, some of the challenges identified previously (see Shafer et al., 2015) continue to limit the application of genomics in plant conservation. At the current state, published or de novo-generated “reference genomes” provide the most easily approached pathway towards the utilizing of genomic data to support the protection of threatened species and environments under threat. Arguments to integrate genomics in the toolbox of conservation biology are well aligned with the vision of major genomics projects such as the “European Reference Genome Atlas” (https://www.erga-biodiversity.eu), the “Darwin Tree of Life” (https://www.darwintreeoflife.org), the “California Conservation Genomics Project” (https://www.ccgproject.org), and the “Earth BioGenome Project” (https://www.earthbiogenome.org). Some of these initiatives such as the “European Reference Genome Atlas” and the “California Conservation Genomics Project” were launched as a response to the threats to biodiversity caused by anthropogenic activities, whereas “Earth BioGenome Project” explicitly states “Sequencing Life for the Future of Life”.
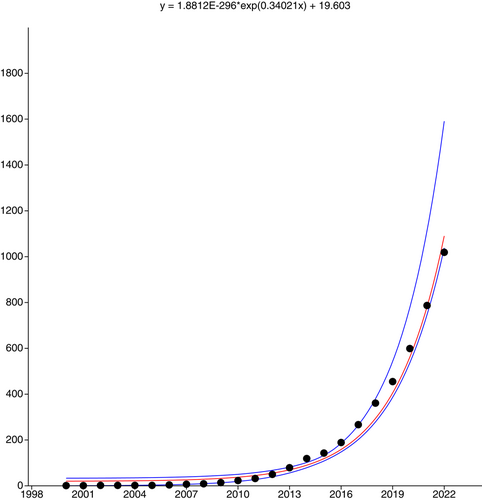
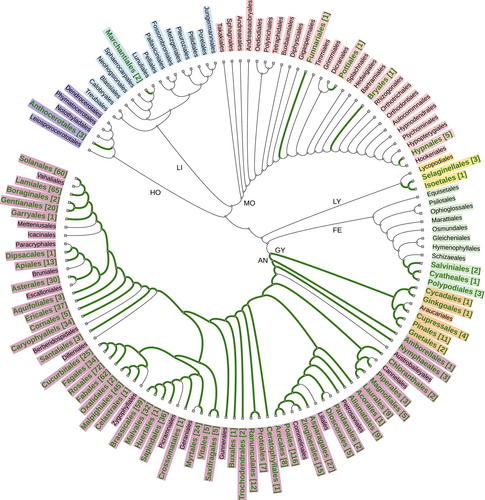
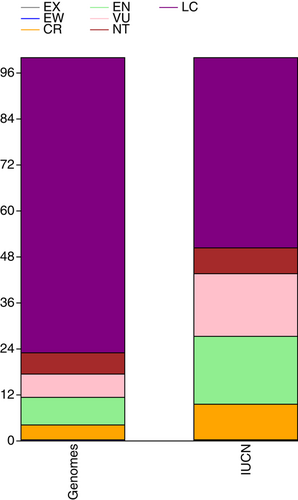
The visionary aims of these initiatives are realistic as the consequence of the amazing progress in sequencing technologies and informatics tools (Pucker et al., 2022). These advances took the genomics of the tree of life into the research realm of biodiversity sciences including conservation biology (Brandies et al., 2019; Formenti et al., 2022; O'Brien 2022; Supple & Shapiro, 2018). In particular, the genomics of the green branch of the tree of life (plants) has seen fast progress since the publication of the first whole-plant genomes in 2000—the genome of the derived angiosperm Arabidopsis thaliana (see Kress et al., 2022; Marks et al., 2021; Szövényi et al., 2021). In September 2022, 1020 plant species, including a few nothospecies, had at least one whole-plant genome published (Supporting Information: Table 1). The current exponential growth of reference genomes (Figure 1) holds the promise that reference genomes will be accessible to all lineages of plants in the near future. Several recent reviewers on plant genomics pointed out the opportunities and challenges such as the crucial role of global participation and attention required to all branches of the green tree of life (e.g., Kress et al., 2022; Marks et al., 2021; Szövényi et al., 2021). Despite a notable bias towards economically important plants and their relatives, recent progress tackled also the black sheep of plant genomics, such as ferns (Kress et al., 2022; Szövényi et al., 2021). Therefore, at least one reference genome is now available for each of the major lineages of land plants (Figure 2). Unfortunately, the current representation still shows a strong bias towards seed plants as reflected by the weak correlation (r2 = 0.997, p (perm.) = 0.079) between the proportional contribution to of each major lineage to the total species richness of land plants and the proportional representation of each lineage to the number of land plant species with at least one genome. In the context of protecting plant diversity, the representation of species according to IUCN Red List Categories revealed a correlation (r2 = 0.896, p (perm.) = 0.002) between the distribution of the categories among plant species evaluated in the IUCN Red Plant List (as available in September 2022) and the distribution among the plant taxa with a published genome sequence and IUCN categorization (Figure 3). This result is notable, although the overlap between the two data sets has been restricted to 361 plant species, corresponding to 35.4% of plant species with a sequence and published genome. As expected, species categorized as Least Concern (77.0%) dominated the list of taxa with a whole genome available while 17.2% of species with a genome sequenced were categorized as threatened. This is remarkable, because conservation needs were rarely considered as a key argument in the selection of species. In turn, access to material suitable for whole-plant genomics may actually limit their utilization.
3 THE PROS AND CONS OF REFERENCE GENOMES
Despite the remarkable rapid increase of fully sequenced plant genomes (see Figure 1), generating complete genome sequences of all land plants will be a far-fetched target for some time to come (Formenti et al., 2022). Fortunately, the application of genomic approaches is not restricted to taxa that have at least one whole genome sequenced. Despite the general utility of the method, the application of genomics to threatened species benefits from access to an accessible reference genome obtained for the same species or phylogenetically close relatives. In short, the utilization of conservation genomics will be much improved by improved coverage of all branches of the tree of life. As visualized in Figure 2, many orders of land plants lack behind because a complete whole genome has not been published for at least one species. Thus, attention given to improving phylogenetic coverage of plant genomes will not only enforce comprehensive analyses of genome architecture but also support conservation genomics of threatened species by improving protein surveys and marker development as the consequence of accurate genome assemblies providing reliable annotation of gene coordinates and gene function. Studies utilizing genomic tools without having access to a reference genome of the same species or a close relative have to take into account the uncertainties caused by the reduced knowledge of genome structure and gene coordinates (Formenti et al., 2022).
4 THE GENETICS OF EXTINCTION—A POORLY UNDERSTOOD PROCESS
In general, highly threatened species are considered to have extremely small effective population sizes as a consequence of recent and rapid population size decline. This decline in the number of individuals contributing to the reproduction of the next generation results in a strong decline in genetic diversity and, in turn, an increase in the frequency of inbreeding. As a consequence, these species show a reduced ability to adapt to changes in their environment, which further enhances the threats to the species. Besides the reduced adaptivity, the highly enhanced inbreeding rate will lead to the accumulation of “negative” mutations that reduce the fitness of the few surviving individuals. The degeneration of the genomes of threatened species has been summarized under the term “genome erosion”, integrating overall homozygosity, genetic load, and runs of homozygosity (Bosse & van Loon, 2022). However, quantification of genome erosion has been recognized to be challenging, although it is crucial to understand the detection of genomic health in species experiencing rapid habitat loss and fragmentation. Furthermore, much may be learned from studying the strategies to reduce the threat of genome erosion that have been explored during the evolution of plants adapted to survive extremely small population sizes located in highly fragmented habitats. For example, many fern species show the ability to extreme inbreeding via gametophytic inbreeding (Sessa et al., 2016). This process produces complete homozygotes that are prone to purification selection (Caballero et al., 2017). Purging genetic load via this reproductive mode provides arguably crucial support to long-distance dispersal events in which the small founder population will experience a high frequency of inbreeding.
At least some plant species have exploited strategies in response to the immanent threat of loss of genetic diversity and genome erosion when experiencing repeated fluctuations of population size, habitat fragmentation, or colonizing new habitats via small founder populations. In fact, very little is known about these strategies but, in turn, we may obtain crucial information by studying these plants to improve the conservation management of highly threatened plant species. The link between maintaining genetic diversity and reproductive modes has been recognized since the introduction of the theoretical framework of population genetics, but new genomic evidence enhances our ability to elucidate the role of these processes in extinction avoidance.
Another closely related research question is focused on maintaining the genomic integrity of species experiencing rapid loss of genetic diversity and putative gene exchange via hybridization with closely related species (Rhymer & Simberloff, 1996). Although introgression with related species may not dramatically change the genetic diversity of species with large and/or stable population sizes, gene exchange with introgression is arguably a major threat to species experiencing rapid population size loss. Their genomes may get swamped by genes from closely related species. Thus, the threatened species may lose their species identity. Until recently, the putative positive aspect of hybrids and introgression has been widely ignored. Some of the genetic diversity may be even maintained in the hybrids formed, where they may be recovered using genomic tools. A recent genomic study on red wolfs in North America confirmed this possibility (vonHoldt et al., 2022). Related evidence was also reported for hybrids formed in the threatened asteroid genus Commidendrum—an endemic to the endangered flora of the Atlantic St. Helena Island (Gray et al., 2017). Thus, the formation of hybrids may have two contrasting aspects. On one hand, introgression threatens the genetic integrity of threatened species but, on the other hand, introgression is a crucial process in genetic rescue as a conservation tool (Ralls et al., 2020; Starkey & Gray, 2022).
5 ACTIONS TOWARD THE INTEGRATION OF GENOMICS INTO PLANT CONSERVATION
To take genomics effectively into the realm of conservation of plant diversity, several initiatives have to be considered such as the Threatened Species Initiatives (Hogg et al., 2022). In the following discussion, I will emphasize two initiatives that set priority to conservation needs in future genome sequencing initiatives. First, prioritizing the sequencing of plant genomes of species known to be threatened at the global or regional scale. These data will provide crucial support to implement effective conservation management strategies. This initiative resembles the “Threatened Species Initiative” proposed by Hogg et al. (2022). Second, obtaining reference genomes of keystone plant species of habitats considered to be under stress as the consequence of environmental changes such as deforestation. These data will not only provide new insights into the ecology and evolution of these habitats but also enable to foster efforts such as re-forestation initiatives.
6 RATIONALE OF INITIATIVE 1—GENERATING REFERENCE GENOMES OF HIGHLY THREATENED SPECIES
Action plans aiming to protect highly threatened species benefit substantially by integrating reference genomes of these species (Brandies et al., 2019; Hogg et al., 2022; Paez et al., 2022; Wang et al., 2022). Species experiencing rapid decline in number of individuals are prone to highly enhanced extinction threats as the consequence of genome erosion (Hogenloge et al., 2020; Bosse & van Loon, 2022; Tian & Ma, 2021). Furthermore, the increased frequency of negative mutations undermines the success of breeding programs aiming to expand the number of individuals, in particular by the enhanced rate of inbreeding. The application of a reference genome enables experimental approaches (Galla et al., 2019) to avoid, or at least reduce, the effect of the previous decline of the genetic pool of a species by establishing carefully designed breeding programs aiming to maximize genetic diversity and limit the accumulation of deleterious mutations. In some cases, these efforts may require genetic rescue, sometimes even using closely related species (Ralls et al., 2020; Whiteley et al., 2015). Reference genomes have now been employed to support the protection of threatened vertebrates via breeding programs or related conservation programs such as the giant panda (Li et al., 2010), the Tasmanian devil (Brandies et al., 2019), Chinese pangolins (Wang et al., 2022), Aldabra giant tortoise (Çilingir et al., 2022), and red wolf (vonHoldt et al., 2022), but have not been widely used in plants until recently. This bias may be partly explained by less promotion of charismatic species such as those animals mentioned above, but this trend is arguably challenged by recent efforts to promote plant conservation by focusing on iconic plants including the living fossil Ginkgo (Liu et al., 2021), palms (Bellot et al., 2022), conifers such as Coast Redwood and Giant sequoia (De La Torre et al., 2021; Neale et al., 2021; Scott et al., 2020), island endemics (Hamabata et al., 2019), and rhododendrons (Ma et al., 2021). Promoting the protection of plant species is arguably the key to raising the resources required to integrate genomics into in situ and ex situ conservation management programs. Some key lessons and technological advances may be taken from the innovative genomic-based breeding technologies employed in our efforts to secure sustainable food production (Anders et al., 2021; Bevan et al., 2017).
Recently, some whole genome sequences were generated with a specific intention to support the conservation of highly endangered plant species such as the eudicot Bretschneidera sinensis (Zhang et al., 2022) and the derived fern Adiantum nelumboides (Zhong et al., 2022). The de novo-generated genomes enable the study of the demographic history of these plants (Sun et al., 2022; Zhang et al., 2022; Zhong et al., 2022) that will be further integrated, together with ecological field surveys (Yu et al., 2022), into the exploration of the protocols of ex situ and in situ conservation measures of these plants.
Genome sequencing of highly endangered species is best promoted by targeting ex situ collections of species classified as highly threatened or even extinct in the wild that are grown at botanical gardens (Abeli et al., 2020; Westwood et al., 2021). These efforts support ongoing ex situ propagating and restoration in the wild programs by enabling genomic tool kits to document genetic diversity that will, in turn, improve the efficiency of the already established programs (Abeli et al., 2020). Similar to efforts taken to revive threatened mammal species, genomic tools may enable the recovery of the genetic integrity and diversity of these species. Having access to genomic information will also support the utility of Urban conservation gardening and other restoration programs (Segar et al., 2022). Finally, genomics will become an integral component of ex situ conservation procedures similar to the already existing gene banking procedures (Supple & Shapiro, 2018). The genomic resources will also be highly valuable to detect unique proteins and pathways producing secondary compounds of which some are of interest to improve food security and/or have medical implications.
7 RATIONALE OF INITIATIVE 2—GENERATING REFERENCE GENOMES OF KEYSTONE SPECIES
Ecosystem restoration has been taken into the center of global efforts towards a healthier planet by the announcement of the United Nations Decade on Ecosystem Restoration (https://www.unesco.org/en/ecosystems-restoration-decade). Whereas the potential of genomics for restoring ecosystems and protecting natural habitats has been recognized, the implementation of genomic approaches has been still a challenge (Breed et al., 2019; Mohr et al., 2022). Obtaining genomes of keystone species such as Vatica mangachapoi—a keystone in a dipterocarp-dominated tropical forest—enabled quantifying local adaptations and genetic load across populations that can be applied to planned pedigree mating and to guide replanting in drought-afflicted areas (Tang et al., 2022). In restoration programs and conservation of threatened species occurring in disjunct ranges, quantification of the genetic variation at the landscape level is crucial to prioritize conservation under scenarios of environmental change (Gougherty et al., 2021).
Efforts to integrate reference genomes in environmental protection are also strongly aligned with community genomic studies focusing on the genomic constitution of keystone species (Schielzeth & Wolf, 2021). The potential of this approach has been demonstrated by the recently published study on the evolution of mangrove habitats utilizing comparative community-wide genomic data—in total 62 species—to elucidate the ecological and evolutionary aspects of a habitat that showed high near-extinction rates in the evolutionary history of these unique environments (Guo et al., 2022; He et al., 2022). In turn, the obtained evidence improved our ability to predict the impact of the anthropogenic enforced habitat loss on the genetic diversity of mangrove taxa in the near future. Determining the mechanism enabling the taxa to survive substantial population size variation under natural climatic fluctuations will not only enable to estimate the thresholds of their tolerance to anthropogenic introduced fragmentation and population size declines but also inform the utilization of genetic rescue and related procedures in protecting these species and restoration of these unique habitats.
Here, I suggest establishing community genomic programs focusing on keystone plant species of putative threatened ecosystems. Specifically, we urgently need a joint initiative using genomic resources to support ongoing efforts to protect the unique plant and animal diversity of the species-rich tropical forests in southern China, such as at the Xishuangbanna Dai Autonomous Prefecture in Southern Yunnan. This initiative will focus on providing genetics to biodiversity protection but also conserve socioeconomic aspects of the natural resources provided by the unique rich biodiversity by the assembly of reference genomes of the key-stone plant species—such as dominant tree species Barringtonia racemosa (L.) Spreng, Lasiococca comberi Haines, Parashorea chinensis H. Wang, Pometia pinnata J.R. Forster & G. Forster, and Tetrameles nudifolora R. Brown (Zhu, 2006), These species are known for their ecological role, socioeconomic importance, and threat status. Besides generating reference genomes of these tree species, attention may also be given to other plants, including species recognized either as natural resources, for example medicinal plants, or of ecological importance. Reference genomes are currently generated for ecologically and socioeconomically important plant species growing in southern Yunnan, such as the gleichenioid fern Dicanopteris pedata (Houtt.) Nakaike—a plant of great interest to ecosystem restoration (Yang et al., 2021)—and the threatened golden chicken fern Cibotium barometz (L.) J. Sm. These plants are of great interest to ecosystem restoration and/or are valued for their medicinal qualities (Howes et al., 2017). As demonstrated by the study on mangrove forests (He et al., 2022), community genomes of rich plant communities are not only achievable but elucidate the evolutionary history of these communities. In turn, the information gained is highly relevant to predict their resistance to rapid reduction and expansion of the space occupied. These insights are crucial in our efforts to maintain the biodiversity of these habitats via the establishment of protected areas as well as efforts to restore ecosystems (Breed et al., 2019).
Initiatives to empower conservation actions by incorporating genomic resources are now widely proposed (see Hogg et al., 2022). Applying genomic approaches to species-rich ecosystems such as tropical rainforest systems take the challenges towards a scale that takes biodiversity conservation towards big data science. In turn, these data will provide crucial support to achieve major targets such as the maintenance of genetic diversity of species considered to be of crucial importance to the maintenance of biodiversity. Furthermore, having access to the genetic blueprint enables us not only to establish ex situ breeding programs designed to maintain genetic diversity but also to identify adaptive traits. Insights provided will not only determine the impact of environmental changes such as global warming on the pool of genetic diversity but also allow the identification of beneficial traits supporting our efforts to restore healthy ecosystems. In some cases, the identified genes will trigger unexpected innovations.
In summary, the rapid progress in plant genomics enabled the accumulation of reference genomes for many but not all orders of land plants. To improve our ability to protect land plant biodiversity and natural resources provided, this study put forward arguments to run initiatives taking plant genomics in the realm of plant conservation.
AUTHOR CONTRIBUTIONS
The sole author designed the study, carried out all analyses, wrote and revised the manuscript.
CONFLICT OF INTEREST STATEMENT
The author declares no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
This manuscript does not include any new data. The data set used to calculate the statistics is provided as a Supporting Information.




