Germline variants of DNA repair and immune genes in lymphoma from lymphoma-cancer families
Xiaogan Wang
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorLijuan Deng
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorLingyan Ping
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorYunfei Shi
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Pathology, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorHaojie Wang
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Central Laboratory, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorFeier Feng
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorXin Leng
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorYahan Tang
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorYan Xie
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorZhitao Ying
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorWeiping Liu
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorJun Zhu
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorCorresponding Author
Yuqin Song
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Correspondence
Yuqin Song, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, No. 52 Fucheng Road, Haidian District, Beijing 100142, China.
Email: [email protected]
Search for more papers by this authorXiaogan Wang
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorLijuan Deng
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorLingyan Ping
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorYunfei Shi
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Pathology, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorHaojie Wang
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Central Laboratory, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorFeier Feng
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorXin Leng
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorYahan Tang
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorYan Xie
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorZhitao Ying
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorWeiping Liu
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorJun Zhu
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Search for more papers by this authorCorresponding Author
Yuqin Song
Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, Beijing, China
Correspondence
Yuqin Song, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Lymphoma, Peking University Cancer Hospital & Institute, No. 52 Fucheng Road, Haidian District, Beijing 100142, China.
Email: [email protected]
Search for more papers by this authorAbstract
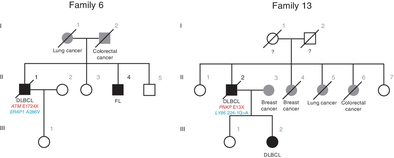
The genetic predisposition to lymphoma is not fully understood. We identified 13 lymphoma-cancer families (2011–2021), in which 27 individuals developed lymphomas and 26 individuals had cancers. Notably, male is the predominant gender in lymphoma patients, whereas female is the predominant gender in cancer patients (p = .019; OR = 4.72, 95% CI, 1.30–14.33). We collected samples from 18 lymphoma patients, and detected germline variants through exome sequencing. We found that germline protein truncating variants (PTVs) were enriched in DNA repair and immune genes. Totally, we identified 31 heterozygous germline mutations (including 12 PTVs) of 25 DNA repair genes and 19 heterozygous germline variants (including 7 PTVs) of 14 immune genes. PTVs of ATM and PNKP were found in two families, respectively. We performed whole genome sequencing of diffuse large B cell lymphomas (DLBCLs), translocations at IGH locus and activation of oncogenes (BCL6 and MYC) were verified, and homologous recombination deficiency was detected. In DLBCLs with germline PTVs of ATM, deletion and insertion in CD58 were further revealed. Thus, in lymphoma-cancer families, we identified germline defects of both DNA repair and immune genes in lymphoma patients.
Graphical Abstract
What's new?
The genetic predisposition to lymphoma has yet to be fully understood. Here, the authors performed sequencing analyses in 13 lymphoma-cancer families, in which males tended to develop lymphomas and females to develop cancers. They identified germline variants for 25 DNA repair genes and 14 immune genes in 18 lymphoma patients, with more than one third of these mutations being protein truncating. Genome instability and oncogene activation were verified in lymphoma samples, and somatic mutations mediating immune evasion were also found. These germline defects of DNA repair and immune genes suggest germline double hits in lymphomagenesis.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The WES, WGS, and RNAseq data generated in this study are available in the National Genomics Data Center (https://ngdc.cncb.ac.cn/omix) under accession numbers OMIX005168 and OMIX005149.41 Other information that supports the findings of this study is available from the corresponding author upon request.
Supporting Information
| Filename | Description |
|---|---|
| ijc34892-sup-0001-Supinfo.pdfPDF document, 1.3 MB | Data S1. Supporting information. |
| ijc34892-sup-0002-Tables.xlsxExcel 2007 spreadsheet , 47 KB | Data S2. Supporting information. |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
REFERENCES
- 1Maldonado N, Levin M. Familial lymphoma: review of the literature and report of two brothers with lymphosarcoma. Bol Asoc Med P R. 1961; 53: 325-337.
- 2Goldin LR, Bjorkholm M, Kristinsson SY, Turesson I, Landgren O. Highly increased familial risks for specific lymphoma subtypes. Br J Haematol. 2009; 146(1): 91-94. doi:10.1111/j.1365-2141.2009.07721.x
- 3Crump C, Sundquist K, Sieh W, Winkleby MA, Sundquist J. Perinatal and family risk factors for non-Hodgkin lymphoma in early life: a Swedish national cohort study. J Natl Cancer Inst. 2012; 104(12): 923-930. doi:10.1093/jnci/djs225
- 4Saarinen S, Pukkala E, Vahteristo P, Makinen MJ, Franssila K, Aaltonen LA. High familial risk in nodular lymphocyte-predominant Hodgkin lymphoma. J Clin Oncol. 2013; 31(7): 938-943. doi:10.1200/JCO.2012.43.5958
- 5Morton LM, Slager SL, Cerhan JR, et al. Etiologic heterogeneity among non-Hodgkin lymphoma subtypes: the InterLymph non-Hodgkin lymphoma subtypes project. J Natl Cancer Inst Monogr. 2014; 2014(48): 130-144. doi:10.1093/jncimonographs/lgu013
- 6Cerhan JR, Slager SL. Familial predisposition and genetic risk factors for lymphoma. Blood. 2015; 126(20): 2265-2273. doi:10.1182/blood-2015-04-537498
- 7Kharazmi E, Fallah M, Pukkala E, et al. Risk of familial classical Hodgkin lymphoma by relationship, histology, age, and sex: a joint study from five Nordic countries. Blood. 2015; 126(17): 1990-1995. doi:10.1182/blood-2015-04-639781
- 8Fallah M, Kharazmi E, Pukkala E, et al. Familial risk of non-Hodgkin lymphoma by sex, relationship, age at diagnosis and histology: a joint study from five Nordic countries. Leukemia. 2016; 30(2): 373-378. doi:10.1038/leu.2015.272
- 9Sud A, Chattopadhyay S, Thomsen H, et al. Analysis of 153 115 patients with hematological malignancies refines the spectrum of familial risk. Blood. 2019; 134(12): 960-969. doi:10.1182/blood.2019001362
- 10Salipante SJ, Mealiffe ME, Wechsler J, et al. Mutations in a gene encoding a midbody kelch protein in familial and sporadic classical Hodgkin lymphoma lead to binucleated cells. Proc Natl Acad Sci U S A. 2009; 106(35): 14920-14925. doi:10.1073/pnas.0904231106
- 11Saarinen S, Aavikko M, Aittomaki K, et al. Exome sequencing reveals germline NPAT mutation as a candidate risk factor for Hodgkin lymphoma. Blood. 2011; 118(3): 493-498. doi:10.1182/blood-2011-03-341560
- 12Rotunno M, McMaster ML, Boland J, et al. Whole exome sequencing in families at high risk for Hodgkin lymphoma: identification of a predisposing mutation in the KDR gene. Haematologica. 2016; 101(7): 853-860. doi:10.3324/haematol.2015.135475
- 13Speedy HE, Kinnersley B, Chubb D, et al. Germline mutations in shelterin complex genes are associated with familial chronic lymphocytic leukemia. Blood. 2016; 128: 2319-2326. doi:10.1182/blood-2016-01-695692
- 14Gayden T, Sepulveda FE, Khuong-Quang DA, et al. Germline HAVCR2 mutations altering TIM-3 characterize subcutaneous panniculitis-like T cell lymphomas with hemophagocytic lymphohistiocytic syndrome. Nat Genet. 2018; 50(12): 1650-1657. doi:10.1038/s41588-018-0251-4
- 15Leeksma OC, de Miranda NF, Veelken H. Germline mutations predisposing to diffuse large B-cell lymphoma. Blood Cancer J. 2017; 7(2):e532. doi:10.1038/bcj.2017.15
- 16Mosquera Orgueira A, Cid Lopez M, Peleteiro Raindo A, et al. Detection of rare germline variants in the genomes of patients with B-cell neoplasms. Cancers (Basel). 2021; 13(6): 1340. doi:10.3390/cancers13061340
- 17Wang Z, Wilson CL, Armstrong GT, et al. Association of Germline BRCA2 mutations with the risk of pediatric or adolescent non-Hodgkin lymphoma. JAMA Oncologia. 2019; 5(9): 1362-1364. doi:10.1001/jamaoncol.2019.2203
- 18Wang X, Song Y, Chen W, et al. Germline variants of DNA repair genes in early onset mantle cell lymphoma. Oncogene. 2021; 40(3): 551-563. doi:10.1038/s41388-020-01542-2
- 19Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010; 141(1): 27-38. doi:10.1016/j.cell.2010.03.016
- 20Gostissa M, Alt FW, Chiarle R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu Rev Immunol. 2011; 29: 319-350. doi:10.1146/annurev-immunol-031210-101329
- 21Helmink BA, Sleckman BP. The response to and repair of RAG-mediated DNA double-strand breaks. Annu Rev Immunol. 2012; 30: 175-202. doi:10.1146/annurev-immunol-030409-101320
- 22Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013; 152(3): 417-429. doi:10.1016/j.cell.2013.01.007
- 23Robbiani DF, Nussenzweig MC. Chromosome translocation, B cell lymphoma, and activation-induced cytidine deaminase. Annu Rev Pathol. 2013; 8: 79-103. doi:10.1146/annurev-pathol-020712-164004
- 24Bednarski JJ, Sleckman BP. At the intersection of DNA damage and immune responses. Nat Rev Immunol. 2019; 19(4): 231-242. doi:10.1038/s41577-019-0135-6
- 25Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009; 25(14): 1754-1760. doi:10.1093/bioinformatics/btp324
- 26Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019; 37(8): 907-915. doi:10.1038/s41587-019-0201-4
- 27Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015; 33(3): 290-295. doi:10.1038/nbt.3122
- 28Scott DW, Gascoyne RD. The tumour microenvironment in B cell lymphomas. Nat Rev Cancer. 2014; 14(8): 517-534. doi:10.1038/nrc3774
- 29Zhu YX, Benn S, Li ZH, et al. The SH3-SAM adaptor HACS1 is up-regulated in B cell activation signaling cascades. J Exp Med. 2004; 200(6): 737-747. doi:10.1084/jem.20031816
- 30von Holleben M, Gohla A, Janssen KP, Iritani BM, Beer-Hammer S. Immunoinhibitory adapter protein Src homology domain 3 lymphocyte protein 2 (SLy2) regulates actin dynamics and B cell spreading. J Biol Chem. 2011; 286(15): 13489-13501. doi:10.1074/jbc.M110.155184
- 31He C, Wang S, Zhou C, et al. CD36 and LC3B initiated autophagy in B cells regulates the humoral immune response. Autophagy. 2021; 17(11): 3577-3591. doi:10.1080/15548627.2021.1885183
- 32Yuan Q, Li Y, Li J, et al. WDFY4 is involved in symptoms of systemic lupus erythematosus by modulating B cell fate via noncanonical autophagy. J Immunol. 2018; 201(9): 2570-2578. doi:10.4049/jimmunol.1800399
- 33Dreyling M, Campo E, Hermine O, et al. Newly diagnosed and relapsed mantle cell lymphoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncology. 2017; 28(Suppl 4): iv62-iv71. doi:10.1093/annonc/mdx223
- 34Cheah CY, Seymour JF, Wang ML. Mantle cell lymphoma. J Clin Oncol. 2016; 34(11): 1256-1269. doi:10.1200/JCO.2015.63.5904
- 35Puente XS, Jares P, Campo E. Chronic lymphocytic leukemia and mantle cell lymphoma: crossroads of genetic and microenvironment interactions. Blood. 2018; 131(21): 2283-2296. doi:10.1182/blood-2017-10-764373
- 36Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021; 71(3): 209-249. doi:10.3322/caac.21660
- 37Lee JH, Paull TT. Cellular functions of the protein kinase ATM and their relevance to human disease. Nat Rev Mol Cell Biol. 2021; 22(12): 796-814. doi:10.1038/s41580-021-00394-2
- 38Bredemeyer AL, Sharma GG, Huang CY, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006; 442(7101): 466-470. doi:10.1038/nature04866
- 39Callen E, Jankovic M, Difilippantonio S, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007; 130(1): 63-75. doi:10.1016/j.cell.2007.06.016
- 40Williams GJ, Hammel M, Radhakrishnan SK, Ramsden D, Lees-Miller SP, Tainer JA. Structural insights into NHEJ: building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Repair (Amst). 2014; 17: 110-120. doi:10.1016/j.dnarep.2014.02.009
- 41 CNCB-NGDC Members and Partners. Database resources of the National Genomics Data Center, China National Center for bioinformation in 2022. Nucleic Acids Res. 2022; 50(D1): D27-D38. doi:10.1093/nar/gkab951