Mutational evolution after chemotherapy-progression in metastatic colorectal cancer revealed by circulating tumor DNA analysis
Sheehyun Kim
Department of Genomic Medicine, Seoul National University Hospital, Seoul, South Korea
Search for more papers by this authorYongjun Cha
Center for Colorectal Cancer, National Cancer Center, Research Institute and Hospital, Goyang, South Korea
Search for more papers by this authorYoojoo Lim
Department of Internal Medicine, Seoul National University Hospital, Seoul, South Korea
Search for more papers by this authorKyung-Hun Lee
Department of Internal Medicine, Seoul National University Hospital, Seoul, South Korea
Cancer Research Institute, Seoul National University, Seoul, South Korea
Search for more papers by this authorMin Jung Kim
Department of Surgery, Seoul National University Hospital, Seoul, South Korea
Search for more papers by this authorJi Won Park
Department of Surgery, Seoul National University Hospital, Seoul, South Korea
Search for more papers by this authorSeung-Bum Ryoo
Department of Surgery, Seoul National University Hospital, Seoul, South Korea
Search for more papers by this authorSeung-Yong Jeong
Department of Surgery, Seoul National University Hospital, Seoul, South Korea
Search for more papers by this authorKyu Joo Park
Department of Surgery, Seoul National University Hospital, Seoul, South Korea
Search for more papers by this authorCorresponding Author
Sae-Won Han
Department of Internal Medicine, Seoul National University Hospital, Seoul, South Korea
Cancer Research Institute, Seoul National University, Seoul, South Korea
Correspondence
Sae-Won Han, Department of Internal Medicine, Seoul National University Hospital, 101 Daehak-ro, Jongno-gu, Seoul, 03080, South Korea.
Email: [email protected]
Search for more papers by this authorTae-You Kim
Department of Internal Medicine, Seoul National University Hospital, Seoul, South Korea
IMBdx, Inc., Seoul, South Korea
Cancer Research Institute, Seoul National University, Seoul, South Korea
Search for more papers by this authorSheehyun Kim
Department of Genomic Medicine, Seoul National University Hospital, Seoul, South Korea
Search for more papers by this authorYongjun Cha
Center for Colorectal Cancer, National Cancer Center, Research Institute and Hospital, Goyang, South Korea
Search for more papers by this authorYoojoo Lim
Department of Internal Medicine, Seoul National University Hospital, Seoul, South Korea
Search for more papers by this authorKyung-Hun Lee
Department of Internal Medicine, Seoul National University Hospital, Seoul, South Korea
Cancer Research Institute, Seoul National University, Seoul, South Korea
Search for more papers by this authorMin Jung Kim
Department of Surgery, Seoul National University Hospital, Seoul, South Korea
Search for more papers by this authorJi Won Park
Department of Surgery, Seoul National University Hospital, Seoul, South Korea
Search for more papers by this authorSeung-Bum Ryoo
Department of Surgery, Seoul National University Hospital, Seoul, South Korea
Search for more papers by this authorSeung-Yong Jeong
Department of Surgery, Seoul National University Hospital, Seoul, South Korea
Search for more papers by this authorKyu Joo Park
Department of Surgery, Seoul National University Hospital, Seoul, South Korea
Search for more papers by this authorCorresponding Author
Sae-Won Han
Department of Internal Medicine, Seoul National University Hospital, Seoul, South Korea
Cancer Research Institute, Seoul National University, Seoul, South Korea
Correspondence
Sae-Won Han, Department of Internal Medicine, Seoul National University Hospital, 101 Daehak-ro, Jongno-gu, Seoul, 03080, South Korea.
Email: [email protected]
Search for more papers by this authorTae-You Kim
Department of Internal Medicine, Seoul National University Hospital, Seoul, South Korea
IMBdx, Inc., Seoul, South Korea
Cancer Research Institute, Seoul National University, Seoul, South Korea
Search for more papers by this authorSheehyun Kim and Yongjun Cha contributed equally to this work.
Abstract
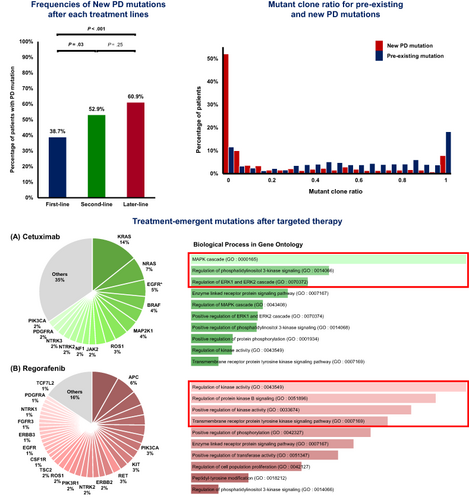
Emerging new mutations after treatment can provide clues to acquired resistant mechanisms. Circulating tumor DNA (ctDNA) sequencing has enabled noninvasive repeated tumor mutational profiling. We aimed to investigate newly emerging mutations in ctDNA after disease progression in metastatic colorectal cancer (mCRC). Blood samples were prospectively collected from mCRC patients receiving palliative chemotherapy before treatment and at radiological evaluations. ctDNA from pretreatment and progressive disease (PD) samples were sequenced with a next-generation sequencing panel targeting 106 genes. A total of 712 samples from 326 patients were analyzed, and 381 pretreatment and PD pairs (163 first-line, 85 second-line and 133 later-line [≥third-line]) were compared. New mutations in PD samples (mean 2.75 mutations/sample) were observed in 49.6% (189/381) of treatments. ctDNA samples from later-line had more baseline mutations (P = .002) and were more likely to have new PD mutations (adjusted odds ratio [OR] 2.27, 95% confidence interval [CI]: 1.40-3.69) compared to first-line. RAS/BRAF wild-type tumors were more likely to develop PD mutations (adjusted OR 1.87, 95% CI: 1.22-2.87), independent of cetuximab treatment. The majority of new PD mutations (68.5%) were minor clones, suggesting an increasing clonal heterogeneity after treatment. Pathways involved by PD mutations differed by the treatment received: MAPK cascade (Gene Ontology [GO]: 0000165) in cetuximab and regulation of kinase activity (GO: 0043549) in regorafenib. The number of mutations revealed by ctDNA sequencing increased during disease progression in mCRC. Clonal heterogeneity increased after chemotherapy progression, and pathways involved were affected by chemotherapy regimens.
Graphical Abstract
What's new?
Mutations that arise during treatment for metastatic colorectal cancer (mCRC) can fuel the development of treatment-resistant tumors. Tracking the mutational evolution of mCRC, however, is challenged by limitations in existing approaches. Here, the authors investigated newly emerging mutations by sequencing circulating tumor DNA (ctDNA) from progressive mCRC patients following conventional treatments. Mutations that emerged in association with treatment were identified in half of progressive disease samples. New mutations were dominantly subclonal variants and were more frequently observed in later-line treatments and RAS/BRAF wild-type tumors. The findings cast new light on relationships between emergent mutations and treatment in progressive mCRC.
CONFLICT OF INTEREST STATEMENT
Yongjun Cha received consulting fees from IMBdx; Yoojoo Lim, Hanseong Roh and Jun-Kyu Kang are employees of IMBdx; Hwang-Phill Kim is an employee and stock owner of IMBdx; Sae-Won Han received honoraria and a research fund from IMBdx and Tae-You Kim is a co-founder and stock owner of IMBdx. The other authors declare no competing interest.
Open Research
DATA AVAILABILITY STATEMENT
All the NGS data of ctDNA sequencing are available on the NCBI Sequence Read Archive (SRA) database with links to BioProject (https://www.ncbi.nlm.nih.gov/bioproject/) accession number PRJNA933526. The data that support the findings of our study are available from the corresponding author upon reasonable request.
Supporting Information
| Filename | Description |
|---|---|
| ijc34558-sup-0001-Supinfo.pdfPDF document, 713.6 KB | Data S1. Supporting Information. |
| ijc34558-sup-0002-TableS2.xlsxExcel 2007 spreadsheet , 67.6 KB | Table S2. Sequencing statistics of study samples. |
| ijc34558-sup-0003-TableS3.xlsxExcel 2007 spreadsheet , 36.7 KB | Table S3. Treatment information with date of blood sampling. |
| ijc34558-sup-0004-TableS4.xlsxExcel 2007 spreadsheet , 68.2 KB | Table S4. List of newly emerging PD mutations. |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
REFERENCES
- 1 National Comprehensive Cancer Network. Colon Cancer. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) 2022; Version 1.2022.
- 2 National Comprehensive Cancer Network. Breast Cancer. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) 2022; Version 4.2022.
- 3 National Comprehensive Cancer Network. Prostate Cancer. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) 2022; Version 4.2022.
- 4National Comprehensive Cancer N. Non-Small Cell Lung Cancer. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®)2022; Version 3.2022.
- 5Mosele F, Remon J, Mateo J, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020; 31: 1491-1505.
- 6Yoon S, Kim M, Hong YS, et al. Recommendations for the use of next-generation sequencing and the molecular tumor board for patients with advanced cancer: a report from KSMO and KCSG precision medicine networking group. Cancer Res Treat. 2022; 54: 1-9.
- 7Westover D, Zugazagoitia J, Cho BC, Lovly CM, Paz-Ares L. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann Oncol. 2018; 29: i10-i19.
- 8Ricordel C, Friboulet L, Facchinetti F, Soria JC. Molecular mechanisms of acquired resistance to third-generation EGFR-TKIs in EGFR T790M-mutant lung cancer. Ann Oncol. 2018; 29: i28-i37.
- 9Leonetti A, Sharma S, Minari R, Perego P, Giovannetti E, Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer. 2019; 121: 725-737.
- 10Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer. 2014; 14: 455-467.
- 11Rizos H, Menzies AM, Pupo GM, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin Cancer Res. 2014; 20: 1965-1977.
- 12Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012; 486: 532-536.
- 13Diaz LA Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012; 486: 537-540.
- 14Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov. 2014; 4: 1269-1280.
- 15Parseghian CM, Loree JM, Morris VK, et al. Anti-EGFR-resistant clones decay exponentially after progression: implications for anti-EGFR re-challenge. Ann Oncol. 2019; 30: 243-249.
- 16Bertotti A, Papp E, Jones S, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015; 526: 263-267.
- 17Bardelli A, Corso S, Bertotti A, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 2013; 3: 658-673.
- 18Troiani T, Martinelli E, Napolitano S, et al. Increased TGF-alpha as a mechanism of acquired resistance to the anti-EGFR inhibitor cetuximab through EGFR-MET interaction and activation of MET signaling in colon cancer cells. Clin Cancer Res. 2013; 19: 6751-6765.
- 19Pietrantonio F, Vernieri C, Siravegna G, et al. Heterogeneity of acquired resistance to anti-EGFR monoclonal antibodies in patients with metastatic colorectal cancer. Clin Cancer Res. 2017; 23: 2414-2422.
- 20Fakih MG, Kopetz S, Kuboki Y, et al. Sotorasib for previously treated colorectal cancers with KRAS(G12C) mutation (CodeBreaK100): a prespecified analysis of a single-arm, phase 2 trial. Lancet Oncol. 2022; 23: 115-124.
- 21Andre T, Shiu KK, Kim TW, et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med. 2020; 383: 2207-2218.
- 22Kopetz S, Grothey A, Yaeger R, et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med. 2019; 381: 1632-1643.
- 23Hong DS, DuBois SG, Kummar S, et al. Larotrectinib in patients with TRK fusion-positive solid tumours: a pooled analysis of three phase 1/2 clinical trials. Lancet Oncol. 2020; 21: 531-540.
- 24Rousseau B, Bieche I, Pasmant E, et al. PD-1 blockade in solid tumors with defects in polymerase epsilon. Cancer Discov. 2022; 12: 1435-1448.
- 25Siena S, Di Bartolomeo M, Raghav K, et al. Trastuzumab deruxtecan (DS-8201) in patients with HER2-expressing metastatic colorectal cancer (DESTINY-CRC01): a multicentre, open-label, phase 2 trial. Lancet Oncol. 2021; 22: 779-789.
- 26Awad MM, Liu S, Rybkin II, et al. Acquired resistance to KRAS(G12C) inhibition in cancer. N Engl J Med. 2021; 384: 2382-2393.
- 27Hazar-Rethinam M, Kleyman M, Han GC, et al. Convergent therapeutic strategies to overcome the heterogeneity of acquired resistance in BRAF(V600E) colorectal cancer. Cancer Discov. 2018; 8: 417-427.
- 28Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019; 20: 71-88.
- 29Alix-Panabieres C, Pantel K. Liquid biopsy: from discovery to clinical application. Cancer Discov. 2021; 11: 858-873.
- 30Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med. 2018; 379: 1754-1765.
- 31Cescon DW, Bratman SV, Chan SM, Siu LL. Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer. 2020; 1: 276-290.
- 32Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol. 2018; 36: 1631-1641.
- 33Cheng ML, Pectasides E, Hanna GJ, Parsons HA, Choudhury AD, Oxnard GR. Circulating tumor DNA in advanced solid tumors: clinical relevance and future directions. CA Cancer J Clin. 2021; 71: 176-190.
- 34Hsu HC, Lapke N, Wang CW, et al. Targeted sequencing of circulating tumor DNA to monitor genetic variants and therapeutic response in metastatic colorectal cancer. Mol Cancer Ther. 2018; 17: 2238-2247.
- 35Leighl NB, Page RD, Raymond VM, et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin Cancer Res. 2019; 25: 4691-4700.
- 36Kim S, Lim Y, Kang JK, et al. Dynamic changes in longitudinal circulating tumour DNA profile during metastatic colorectal cancer treatment. Br J Cancer. 2022; 127: 898-907.
- 37Lee D-W, Han S-W, Lim Y, et al. Abstract 5157: ctDNA change predicts treatment outcome of regorafenib in metastatic colorectal cancer. Cancer Res. 2022; 82: 5157. doi:10.4143/crt.2023.268
10.1158/1538-7445.AM2022-5157 Google Scholar
- 38Meddeb R, Pisareva E, Thierry AR. Guidelines for the preanalytical conditions for analyzing circulating cell-free DNA. Clin Chem. 2019; 65: 623-633.
- 39Salvianti F, Gelmini S, Costanza F, et al. The pre-analytical phase of the liquid biopsy. New Biotechnol. 2020; 55: 19-29.
- 40Volckmar AL, Sultmann H, Riediger A, et al. A field guide for cancer diagnostics using cell-free DNA: from principles to practice and clinical applications. Genes Chromosomes Cancer. 2018; 57: 123-139.
- 41Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN]. 2013. https://github.com/lh3/bwa
- 42Chen S, Zhou Y, Chen Y, et al. Gencore: an efficient tool to generate consensus reads for error suppressing and duplicate removing of NGS data. BMC Bioinform. 2019; 20: 606.
- 43Lai Z, Markovets A, Ahdesmaki M, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016; 44:e108.
- 44Breiman L. Random forests. Mach Learn. 2001; 45: 5-32.
- 45Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013; 14: 128.
- 46Normanno N, Esposito Abate R, Lambiase M, et al. RAS testing of liquid biopsy correlates with the outcome of metastatic colorectal cancer patients treated with first-line FOLFIRI plus cetuximab in the CAPRI-GOIM trial. Ann Oncol. 2018; 29: 112-118.
- 47Lim Y, Kim S, Kang JK, et al. Circulating tumor DNA sequencing in colorectal cancer patients treated with first-line chemotherapy with anti-EGFR. Sci Rep. 2021; 11: 16333.
- 48Wang F, Huang YS, Wu HX, et al. Genomic temporal heterogeneity of circulating tumour DNA in unresectable metastatic colorectal cancer under first-line treatment. Gut. 2022; 71: 1340-1349.
- 49Gupta R, Othman T, Chen C, Sandhu J, Ouyang C, Fakih M. Guardant360 circulating tumor DNA assay is concordant with FoundationOne next-generation sequencing in detecting actionable driver mutations in anti-EGFR naive metastatic colorectal cancer. Oncologist. 2020; 25: 235-243.
- 50Van't ERVE I, Greuter MJE, Bolhuis K, et al. Diagnostic strategies toward clinical implementation of liquid biopsy RAS/BRAF circulating tumor DNA analyses in patients with metastatic colorectal cancer. J Mol Diagn. 2020; 22: 1430-1437.
- 51Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015; 21: 795-801.
- 52Max Ma X, Bendell JC, Hurwitz HI, et al. Disease monitoring using post-induction circulating tumor DNA analysis following first-line therapy in patients with metastatic colorectal cancer. Clin Cancer Res. 2020; 26: 4010-4017.
- 53Kasi PM, Budde G, Krainock M, et al. Circulating tumor DNA (ctDNA) serial analysis during progression on PD-1 blockade and later CTLA-4 rescue in patients with mismatch repair deficient metastatic colorectal cancer. J Immunother Cancer. 2022; 10: 10.
- 54Gouda MA, Huang HJ, Piha-Paul SA, et al. Longitudinal monitoring of circulating tumor DNA to predict treatment outcomes in advanced cancers. JCO Precis Oncol. 2022; 6:e2100512.
- 55Strickler JH, Loree JM, Ahronian LG, et al. Genomic landscape of cell-free DNA in patients with colorectal cancer. Cancer Discov. 2018; 8: 164-173.
- 56Ni Leathlobhair M, Lenski RE. Population genetics of clonally transmissible cancers. Nat Ecol Evol. 2022; 6: 1077-1089.
- 57Raghav K, Ou FS, Venook AP, et al. Acquired genomic alterations on first-line chemotherapy with cetuximab in advanced colorectal cancer: circulating tumor DNA analysis of the CALGB/SWOG-80405 trial (Alliance). J Clin Oncol. 2022; 41: 472-478.
- 58Topham JT, O'Callaghan CJ, Feilotter H, et al. Circulating tumor DNA identifies diverse landscape of acquired resistance to anti-epidermal growth factor receptor therapy in metastatic colorectal cancer. J Clin Oncol. 2022; 41: 485-496.
- 59Kim TW, Peeters M, Thomas A, et al. Impact of emergent circulating tumor DNA RAS mutation in panitumumab-treated chemoresistant metastatic colorectal cancer. Clin Cancer Res. 2018; 24: 5602-5609.
- 60Sartore-Bianchi A, Pietrantonio F, Lonardi S, et al. Circulating tumor DNA to guide rechallenge with panitumumab in metastatic colorectal cancer: the phase 2 CHRONOS trial. Nat Med. 2022; 28: 1612-1618.
- 61 Heinemann PDDmV. A randomised study to assess the efficacy of cetuximab rechallenge in patients with metastatic colorectal cancer (RAS wild-type) responding to first-line treatment with FOLFIRI plus cetuximab. clinicaltrials.gov 2021.
- 62Nakajima H, Kotani D, Bando H, et al. REMARRY and PURSUIT trials: liquid biopsy-guided rechallenge with anti-epidermal growth factor receptor (EGFR) therapy with panitumumab plus irinotecan for patients with plasma RAS wild-type metastatic colorectal cancer. BMC Cancer. 2021; 21: 674.
- 63Morelli MP, Overman MJ, Dasari A, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol. 2015; 26: 731-736.