Cas9-guided haplotyping of three truncation variants in autosomal recessive disease
Abstract
An autosomal recessive disease is caused by biallelic loss-of-function mutations. However, when more than two disease-causing variants are found in a patient's gene, it is challenging to determine which two of the variants are responsible for the disease phenotype. Here, to decipher the pathogenic variants by precise haplotyping, we applied nanopore Cas9-targeted sequencing (nCATS) to three truncation COL7A1 variants detected in a patient with recessive dystrophic epidermolysis bullosa (EB). The distance between the most 5′ and 3′ variants was approximately 19 kb at the level of genomic DNA. nCATS successfully demonstrated that the most 5′ and 3′ variants were located in one allele while the variant in between was located in the other allele. Interestingly, the proband's mother, who was phenotypically intact, was heterozygous for the allele that harbored the two truncation variants, which could otherwise be misinterpreted as those of typical recessive dystrophic EB. Our study highlights the usefulness of nCATS as a tool to determine haplotypes of complicated genetic cases. Haplotyping of multiple variants in a gene can determine which variant should be therapeutically targeted when nucleotide-specific gene therapy is applied.
Biallelic mutations are required for the development of autosomal recessive diseases. Generally, each disease-causing variant is present in each allele. In contrast, it is exceedingly rare to detect three or more variants in a causative gene in a patient. In such a case, determining which two variants are disease-causing has proved challenging because of the inability to discern haplotypes using conventional sequencing technologies.
Single-molecule third-generation sequencing systems (e.g., the nanopore sequencers of Oxford Nanopore Technologies and the PacBio sequencers) can be pivotal in this field. These sequencers typically generate long to ultra-long reads. A combination of CRISPR/Cas9 technology and nanopore sequencing has been further shown to enable enrichment and sequencing of specific regions in genomic DNA (gDNA) (Gilpatrick et al., 2020) without PCR procedures or reverse transcription procedures, which can induce artificial homologous recombination (Meyerhans et al., 1990; Natsuga et al., 2022; Negroni et al., 1995). This method is called nanopore Cas9-targeted sequencing (nCATS) and has been utilized to detect somatic mutations and chromosomal rearrangements (Gilpatrick et al., 2020; Natsuga et al., 2022; Stangl et al., 2020; Watson et al., 2020; Wongsurawat et al., 2020).
Here, by taking advantage of three truncation COL7A1 variants found in a recessive dystrophic epidermolysis bullosa (EB) patient, we show that nCATS can delineate the haplotypes of multiple variants, thus deciphering the disease-causing ones.
The proband was born to nonconsanguineous parents and suffered from EB, a congenital skin fragility disorder (Has et al., 2020), as indicated by skin erosions and blisters at birth (Figure 1a). His EB subtype was classified as recessive dystrophic EB (Has et al., 2020), based on the following factors: (1) the absence of anchoring fibrils on electron microscopy (Figure 1b); (2) the absence of Type VII collagen (COL7), which is the main component of anchoring fibrils (Watanabe et al., 2018), at the dermo-epidermal junction; and (3) skin-split at the level beneath laminin 332 (L332) and Type IV collagen (COL4), which are the components of the basement membrane (lamina densa) (Figure 1c).
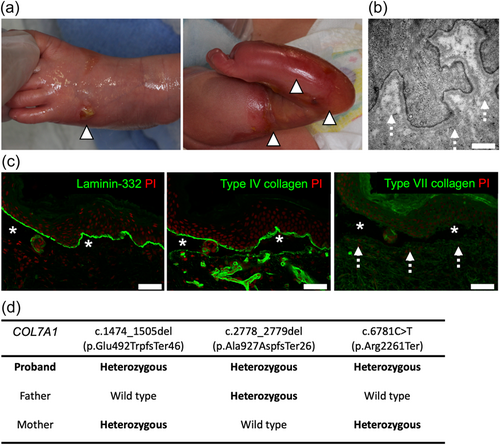
Whole-exome sequencing and subsequent Sanger sequencing of the proband's gDNA revealed three heterozygous truncation variants in COL7A1 (NM000094.4), which encodes COL7 (Figure 1d and Figure S1). All variants were expected to lead to premature termination codons. c.1474_1505del (p.Glu492TrpfsTer46) and c.2778_2779del (p.Ala927AspfsTer26) were novel COL7A1 variants, while c.6781C > T (p.Arg2261Ter; dbSNP, rs772381373) was previously described in RDEB cases (Kern et al., 2006, 2009; Sato-Matsumura et al., 2003).
The mutational profile of the proband indicated the three possible diplotypes, in which two of the three variants were present in one allele and the remaining variant was present in the other allele (diplotype-1, -2, and -3 in Figure 2a). We reasoned that nCATS (Gilpatrick et al., 2020) on the proband's gDNA would precisely identify the haplotypes allocated by the variants. We designed three nCATS experiments (nCATS-1, nCATS-2, and nCATS-3 in Figure 2a), each of which spanned either two of the three or all the variants. The length of the reads obtained by each nCATS experiment was 4, 15, and 19 kb, respectively. The coverage of the pooled samples was 87k (0.9 Gb). nCATS-1 primarily detected two kinds of reads, each harboring c.1475_1505del or c.2778_2779del (Figure 2b and Table S1). Most of the reads of nCATS-2, covering c.2778_2779del and c.6781C > T, also had one of the two variants, but not both (Figure 2c and Table S2). In line with these results, nCATS-3 identified the reads with both c.1474_1505del and c.6781C > T. The other major reads harbored only c.2778_2779del (Figure 2d and Table S3). These results demonstrated that the most 5′ and 3′ variants were present in one allele, and the variant in between was present in the other allele (Proband, Figure 2e).
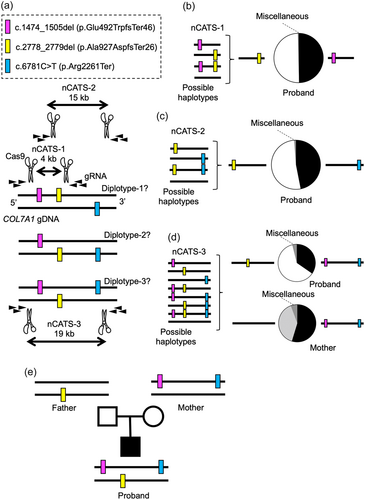
To determine whether the variants of the proband were inherited from the parents, who were phenotypically intact, or developed de novo, we performed Sanger sequencing on the parents' gDNA. The father was heterozygous for c.2778_2779del while the mother was heterozygous for c.1474_1505del and c.6781C > T (Figure 1d and Figure S1). We further confirmed that the mother's c.1474_1505del and c.6781C > T were in the same allele by nCATS-3 (coverage: 56k [0.5 Gb]) (Figure 2d and Table S4). These data revealed that, of the three truncation variants of the proband, one was inherited from the father while the other two came from the mother (Figure 2e).
Our data suggest that c.1474_1505del and c.2778_2779del are mainly pathogenic for recessive dystrophic EB in the proband because c.1474_1505del is located upstream of c.6781C > T in the same allele. This diplotype information also dictates that, when nucleotide-specific gene therapy (e.g., gene editing) is applied, c.2778_2779del should be targeted. By contrast, correction of one of c.1474_1505del or c.6781C > T would be insufficient because the other variant would still be present in the same allele. Notably, the mother harbored two heterozygous truncation mutations. Moreover, her Sanger sequencing results could have been interpreted as those of typical recessive dystrophic EB if haplotyping by nCATS had not been performed.
Although regular genetic testing might not always need nCATS, this method can be an option for haplotyping somatic mutations (Natsuga et al., 2022), haplotyping three or more variants in one patient like our study, and identifying repeat expansions (Mizuguchi et al., 2021; Sone et al., 2019). One nCATS experiment costs hundreds of USD, mostly for a MinION flow cell. Flongle, a smaller flow cell, might reduce the cost if its use is verified for nCATS in the future.
In our study, only nCATS-3 was ultimately sufficient to determine the proband's diplotype. However, as the reads of nCATS-3 were much fewer than those of nCATS-1 and nCATS-2 (Table S1–S4), nCATS-1 and nCATS-2 were confirmatory for this analysis. Furthermore, in scenarios where the most 5' and 3' variants are far from each other (e.g., >50 kb), the combination of nCATS experiments (like nCATS-1 and nCATS-2 in our study) will be needed.
In summary, our study has shown that nCATS is a promising tool for deciphering pathogenic mutations among multiple variants by precise haplotyping. Thus, the haplotypes identified by nCATS can help clinicians design the optimal gene therapy for patients.
ACKNOWLEDGEMENTS
We thank Ms. Mika Tanabe for her technical assistance. This work was supported by the World-leading Innovative and Smart Education (WISE) Program (1801) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, and AMED (ID: 21ak0101168h0001).
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
All sequence data produced by MinION sequencers and haplotype count data were deposited to Gene Expression Omnibus (GEO) with accession no. GSE196673.




