Combined in vitro and in silico analyses of missense mutations in GNPTAB provide new insights into the molecular bases of mucolipidosis II and III alpha/beta
Abstract
Mucolipidosis (ML) II and III alpha/beta are inherited lysosomal storage disorders caused by mutations in GNPTAB encoding the α/β-precursor of GlcNAc-1-phosphotransferase. This enzyme catalyzes the initial step in the modification of more than 70 lysosomal enzymes with mannose 6-phosphate residues to ensure their intracellular targeting to lysosomes. The so-called stealth domains in the α- and β-subunit of GlcNAc-1-phosphotransferase were thought to be involved in substrate recognition and/or catalysis. Here, we performed in silico alignment analysis of stealth domain-containing phosphotransferases and showed that the amino acid residues Glu389, Asp408, His956, and Arg986 are highly conserved between different phosphotransferases. Interestingly, mutations in these residues were identified in patients with MLII and MLIII alpha/beta. To further support the in silico findings, we also provide experimental data demonstrating that these four amino acid residues are strictly required for GlcNAc-1-phosphotransferase activity and thus may be directly involved in the enzymatic catalysis.
Mucolipidosis (ML) type II (MIM# 252500) and type III alpha/beta (MIM# 252600) are autosomal recessive lysosomal storage disorders of childhood (Cathey et al., 2008; Cathey et al., 2010). Both diseases are caused by mutations in the GNPTAB gene encoding the α/β-precursor of N-acetylglucosamine (GlcNAc)-1-phosphotransferase (EC 2.7.8.17; Tiede et al., 2005). This enzyme complex is required for the formation of mannose 6-phosphate (M6P) residues on lysosomal enzymes, which is essential for their intracellular trafficking to lysosomes (Braulke & Bonifacino, 2009). Cells from patients with MLII and MLIII alpha/beta are biochemically characterized by missorting and hypersecretion of lysosomal enzymes into the extracellular space. The subsequent intracellular deficiencies of these lysosomal enzymes result in the accumulation of nondegraded material in dysfunctional lysosomes, which strongly impairs cellular function and homeostasis (Kollmann et al., 2010). Clinical features of MLII-affected individuals include facial dysmorphism, growth retardation, severe skeletal abnormalities, progressive psychomotor retardation, organomegaly, and cardiorespiratory insufficiency leading to death in early childhood. MLIII alpha/beta is an attenuated form of the disease, with later onset and slower progression of the clinical course, which enables survival into adulthood. So far, 258 mutations in the GNPTAB gene have been identified in individuals with MLII and MLIII alpha/beta (Velho et al., 2019). Frameshift and nonsense mutations on both alleles are associated with absent or strongly reduced GlcNAc-1-phosphotransferase activity and result generally in the severe MLII disease. Conversely, missense GNPTAB mutations are mainly associated with residual GlcNAc-1-phosphotransferase activity and result in the MLIII alpha/beta disease with less severe progression (Cathey et al., 2010). Nevertheless, approximately 40% of all identified GNPTAB missense mutations were found in severely affected patients with MLII (Velho et al., 2019).
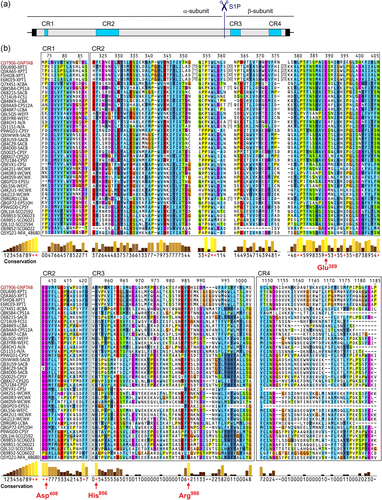
The GlcNAc-1-phosphotransferase is a hexameric complex consisting of two α-, two β-, and two γ-subunits (Bao, Elmendorf, Booth, Drake, & Canfield, 1996). The α- and β-subunits that are synthesized in the endoplasmic reticulum (ER) as common α/β-precursor membrane protein assemble with the soluble γ-subunits to an inactive complex. Cytosolic sorting motifs of the α/β-precursor mediate its subsequent transport from the ER to the Golgi apparatus (Franke, Braulke, & Storch, 2013). In the Golgi apparatus, the α/β-precursor is proteolytically cleaved between Lys928 and Asp929 by site-1 protease (S1P) into the enzymatically active α- and β-subunits (Figure 1a; Marschner, Kollmann, Schweizer, Braulke, & Pohl, 2011). We and others have previously shown that binding of the γ-subunit to the α-subunit enhances the GlcNAc-1-phosphotransferase activity for M6P modification of specific lysosomal enzymes (De Pace et al., 2015; Di Lorenzo et al., 2018; Qian et al., 2010).
For catalytic reaction, the GlcNAc-1-phosphotransferase complex has to exhibit binding sites for the substrates UDP-GlcNAc (phosphate donor) and lysosomal enzymes (phosphate acceptors). Following the transfer of GlcNAc-1-phosphate from the UDP-GlcNAc to the high-mannose-type oligosaccharides on lysosomal enzymes, an α-N-acetylglucosaminidase catalyzes the hydrolysis of the GlcNAc residues, thus exposing the M6P signal (Pohl, Marschner, Storch, & Braulke, 2009; Reitman, Varki, & Kornfeld, 1981).
The binding site for lysosomal enzymes was found to be located in the luminal Notch repeat-like domains and DNA methyltransferase-associated protein domain of the α-subunit (Qian et al., 2015; Qian, Flanagan-Steet, van Meel, Steet, & Kornfeld, 2013; van Meel et al., 2016). The α- and the β-subunit both contain two so-called stealth conserved regions: CR1 and CR2 in the α-subunit and CR3 and CR4 in the β-subunit (Figure 1a). Stealth domains were found in a number of bacterial hexose-1-phosphotransferases involved in the biosynthesis of α-1,4-linked GlcNAc-1-phosphate in capsule polysaccharides (Muindi et al., 2014; Sperisen, Schmid, Bucher, & Zilian, 2005; Tzeng, Noble, & Stephens, 2003), suggesting that the stealth domains harbor the UDP-GlcNAc binding site. Interestingly, amino acid substitutions in the stealth domains of GlcNAc-1-phosphotransferase represent the majority of all missense mutations in patients with MLII and MLIII alpha/beta (Velho et al., 2019). Functional analyses of selected GNPTAB missense mutations in the stealth domains revealed reduced or absent GlcNAc-1-phosphotransferase activity, which was not caused by impaired ER-Golgi transport and/or proteolytic activation of the α/β-precursor (De Pace et al., 2014; Ludwig et al., 2017; Qian et al., 2015; Velho et al., 2015).
To analyze the role of the stealth conserved regions for catalytic activity of GlcNAc-1-phosphotransferase in more detail, first we selected 40 members of the phosphotransferase family from yeast and bacteria, which comprise at least one stealth conserved region (Figure 1b; Table S1), and performed alignment of their amino acid sequences against each of the known stealth conserved regions within the GlcNAc-1-phosphotransferase α/β-precursor. We identified a number of amino acid residues that are fully or highly conserved between the given phosphotransferases, including Pro355, Trp357, Leu358, Leu380, Pro381, Glu389, Ile392/Leu, Ile395/Leu, Ile/Leu398, the Asn406-Asp407-Asp408 motif, and Phe420 in the CR2 domain as well as His956 and Arg986 in the CR3 domain (Figure 1b, indicated by red asterisks or plus signs below the alignment). Whereas the Trp357, Leu358, Leu380, Ile392, Ile395, Leu398, and Phe420 residues in the CR2 domain may be crucial for intermolecular and/or intramolecular hydrophobic interactions of GlcNAc-1-phosphotransferase, the conformation of Pro355 and Pro381 may stabilize the molecular structure of the enzyme and, in particular, of the CR2 domain (Dill, 1990). Most importantly, among conserved residues within the stealth domains, missense mutations leading to amino acid substitutions of Glu389, Asp408, His956, and Arg986 have been identified in individuals affected by MLII and MLIII alpha/beta, that is, p.Glu389Lys (Velho et al., 2019), p.Asp408Asn (Wang et al., 2018), p.His956Tyr (Otomo et al., 2009), p.His956Arg (Cathey et al., 2010), p.Arg986Cys (Cobos, Steglich, Santer, Lukacs, & Gal, 2015; Coutinho et al., 2012), p.Arg986Gly (Velho et al., 2019), and p.Arg986His (Mistri et al., 2018). The clinical description and the molecular analysis of the patient with severe MLII harboring the homozygous mutation p.Glu389Lys (Velho et al., 2019) have become available and are presented here for the first time (Supporting Information). In addition, all used in silico pathogenicity prediction programs, including CADD, REVEL, and M-CAP (Ioannidis et al., 2016; Jagadeesh et al., 2016; Kircher et al., 2014), predict each of the abovementioned variants to have a deleterious effect (Table S2). The data suggest an essential role of these amino acids for the function of GlcNAc-1-phosphotransferase.
Amino acid substitutions in the α/β-precursor may impair the transport of the newly synthesized α/β-precursor protein from the ER to the Golgi apparatus, the proteolytic activation by S1P, or the substrate binding and subsequent catalytic reaction of GlcNAc-1-phosphotransferase. Therefore, to assess molecular consequences of GNPTAB missense mutations, a comprehensive functional analysis of the mutant enzyme is necessary. Whereas the missense mutations p.His956Tyr and p.Arg986Cys have been analyzed in previous studies (De Pace et al., 2014; Qian et al., 2015; Velho et al., 2019), functional analyses of the recently identified mutations p.Glu389Lys and p.Asp408Asn have been performed for the first time in this study. To adequately support the hypothesis that we raised based on the in silico sequence alignment, the in vitro data obtained for all the four missense mutations are shown side by side for better presentation (Figure 2). Briefly (for details, see the online Supporting Information), the missense mutations were individually introduced into the wild-type (WT) complementary DNA of a human α/β-precursor construct by site-directed mutagenesis (Figure 2a). For the analysis of the subcellular localization of the mutants, we performed immunofluorescence microscopy of transfected HeLa cells using a generated monoclonal antibody against the α-subunit (De Pace et al., 2014) and organelle-specific marker proteins. In agreement with previously published data, the WT α/β-precursor as well as the p.His956Tyr and p.Arg986Cys mutants were found to colocalize with the cis-Golgi marker protein GM130 (Figure 2b; De Pace et al., 2014; Qian et al., 2015). Similarly, the hitherto noncharacterized p.Glu389Lys and p.Asp408Asn mutants can also exit the ER and reach the cis-Golgi apparatus (Figure 2b). Of note, a portion of both p.Glu389Lys and p.Asp408Asn mutant proteins was localized in the ER, as revealed by costaining with the ER marker protein disulfide isomerase (Figure 2b). In line with the localization of the mutants in the cis-Golgi apparatus, 190-kDa α/β-precursors and 45-kDa β-subunits were detectable in HEK-293 cell extracts overexpressing the WT or the mutant proteins (Figure 2c), demonstrating proper cleavage by S1P required for GlcNAc-1-phosphotransferase activity. Finally, the in vitro GlcNAc-1-phosphotransferase activity was measured in extracts of HEK-293 cells overexpressing the WT or the mutant α/β-precursors using [3H]UDP-GlcNAc and α-methylmannoside as phosphate donor and acceptor, respectively (Qian et al., 2015; Velho et al., 2015). All the mutants exhibited less than 5% of WT GlcNAc-1-phosphotransferase activity (Figure 2d). In summary, these data clearly demonstrate that amino acid substitutions of the conserved Glu389, Asp408, His956, and Arg986 residues do not abolish the intracellular transport to the Golgi apparatus and the subsequent S1P-mediated cleavage of the α/β-precursors, but they do lead to a drastic reduction of the GlcNAc-1-phosphotransferase activity.
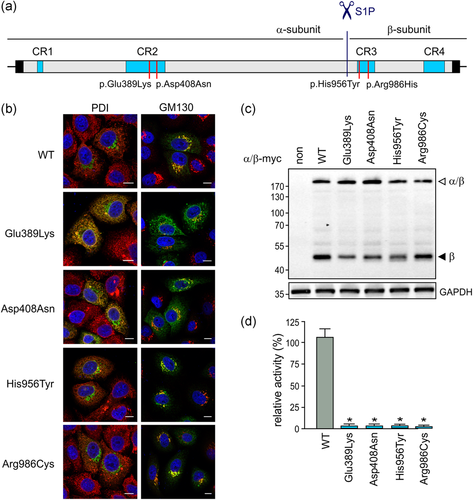
Based on the previously obtained and newly generated data, we hypothesize that the conserved Glu389, Asp408, His956, and Arg986 residues, which are located in the stealth domains CR2 and CR3 of GlcNAc-1-phosphotransferase and are related to MLII and MLIII alpha/beta, play a pivotal role in the UDP-GlcNAc binding and/or may be directly involved in the GlcNAc-1-phosphotransferase-mediated catalysis. It has previously been published that the mutant GlcNAc-1-phosphotransferase α/β-precursor that does not undergo cleavage by S1P is inactive (Marschner et al., 2011), indicating that both α- and β-subunits mediate catalytic function of the active complex. Intriguingly, the identified conserved residues in GlcNAc-1-phosphotransferase are located on the opposite sides of the S1P cleavage site; Glu389 and Asp408 reside in the CR2 domain of the α-subunit whereas His956 and Arg986 are found in the CR3 domain of the β-subunit (Figure 2a). It is thus tempting to speculate that following S1P cleavage of the GlcNAc-1-phosphotransferase α/β-precursor and assembly of the active complex, these four residues may appear proximate enough to drive catalysis. According to the EzCatDB database (http://ezcatdb.cbrc.jp), residues that are most frequently found to be catalytic in transferases responsible for the transfer of nucleotidyl and phosphorous-containing groups include arginine, lysine, aspartic acid and, to a lesser extent, histidine and serine. In addition, aspartic and glutamic acid residues may also play a role in binding divalent ions, such as Mg2+ and Mn2+, which are strictly required for GlcNAc-1-phosphotransferase activity (Bao et al., 1996). Although the conserved residues Glu389, Asp408, His956, and Arg986 are likely directly involved in GlcNAc-1-phosphotransferase catalysis, the exact mechanism of the catalytic reaction mediated by GlcNAc-1-phosphotransferase remains unknown due to lack of data on the tertiary structure of the enzyme. Therefore, further in-depth investigation of the enzyme catalysis demands structural analysis of either the whole GlcNAc-1-phosphotransferase protein complex or other stealth domain-containing phosphotransferases, as knowledge of the structure and biochemistry of these enzymes will help us to better understand the pathogenesis of MLII and MLIII.
ACKNOWLEDGMENTS
The authors are grateful to the patient and her parents for their support in this study. The authors would also like to thank Inka Jantke, Johannes Brand, and Elif Yilmaz for excellent technical assistance, Kerstin Kutsche for financial support as well as the Microscopy Imaging Facility and the Isotope Lab Facility of the University Medical Center Hamburg-Eppendorf for technical support. This study was funded by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, SFB877-B3, PO 1539/1-1, KU 1240/10-1) and the Brazilian National Council for Scientific and Technological Development (CNPq).
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
ETHICS STATEMENT
All procedures carried out with human subjects were in compliance with the Helsinki Declaration. The clinical data presented in this study were retrieved from the routine clinical care facilities of Cerrahpasa School of Medicine, Istanbul, Turkey. Written informed consent was obtained from the parents of the affected child.
Open Research
DATA AVAILABILITY STATEMENT
All mentioned mutations are included in the Leiden Open Variant Database (Fokkema et al., 2011): https://databases.lovd.nl/shared/configuration/GNPTAB. All raw data are available upon request.