Association analysis of exome variants and refraction, axial length, and corneal curvature in a European–American population
Communicated by Peter J. Oefner
Abstract
Refractive errors, myopia, and hyperopia are common visual disorders greatly affecting older individuals. Refraction is determined by genetic factors but only a small percentage of its variation has been explained. We performed a genetic association analysis with three ocular phenotypes: spherical equivalent (a continous measure of refraction), axial length, and corneal curvature in 1,871 European–Americans from the Beaver Dam Eye Study. Individuals were genotyped on the Illumina exome array and imputed to the Haplotype Reference Consortium reference panel. After increasing the number of analyzed variants in targeted protein-coding regions 10-fold via imputation, we confirmed associations for two previously known loci with corneal curvature (chr4q12, rs2114039; g.55092626T > C, β = −0.03 (95% confidence interval [CI]): −0.06, −0.01, P value = 0.01) and spherical equivalent (chr15q14, rs634990; g.35006073T > C, β = −0.27, 95% CI: −0.45, −0.09, P value = 3.79 × 10−3). Despite increased single nucleotide polymorphism (SNP) density, we did not detect any novel significant variants after correction for multiple comparisons. In summary, we confirmed two previous loci associated with corneal curvature and spherical equivalent in a European–American population highlighting the potential biological role of those regions in these traits.
1 INTRODUCTION
Refractive errors, myopia (nearsightedness), and hyperopia (farsightedness) are the most common visual disorders in the world resulting in blurred vision (Centers for Disease Control and Prevention, 2013; Hysi, Wojciechowski, Rahi, & Hammond, 2014; Meng, Butterworth, Malecaze, & Calvas, 2011. Older individuals are disproportionately affected (Resnikoff, Pascolini, Mariotti, & Pokharel, 2008). Approximately only 3.4% of the genetic variation in refraction has been accounted for to date, leaving a significant proportion of “missing heritability” yet to be explained (Hysi et al., 2014; Wojciechowski, 2011). Axial length and corneal curvature are primary biological determinants of refraction, and all three traits are highly heritable with shared etiologies (Biino et al., 2005; Chen et al., 2007, 2012, 2014; Dirani et al., 2006; Dirani, Islam, Shekar, & Baird, 2008; Dirani, Shekar, & Baird, 2008; Guggenheim, McMahon et al., 2013; Guggenheim, Zhou et al., 2013; Hammond, Snieder, Gilbert, & Spector, 2001; Klein et al., 2009; Lyhne, Sjølie, Kyvik, & Green, 2001; Meng et al., 2011; Paget, Vitezica, Malecaze, & Calvas, 2008; Teikari, O'Donnell, Kaprio, & Koskenvuo, 1989; Valluri et al., 1999; Young, Metlapally, & Shay, 2007). Identifying variants associated with axial length and corneal curvature in addition to refraction could account for a proportion of the “missing heritability” and enhance our understanding of the genetic architecture of refraction.
Genotype imputation provides an efficient statistical method for inferring genotypes at variant sites not available on existing arrays, including genome-wide arrays containing common variants (minor allele frequency [MAF] > 0.05) (Das et al., 2016; Delaneau, Marchini, & Zagury, 2012; Delaneau, Zagury, & Marchini, 2013) and exome arrays consisting primarily of rare (MAF < 0.01) and low-frequency (0.01 ≤ MAF ≤ 0.05) variants in selectively targeted exons. Even though imputation from exome arrays has historically been less successful than genome arrays (Kim, Lee, Kim, Consortium, & Park, 2015; Martin, Tse, Bustamante, & Kenny, 2014; Nelson et al., 2013; Pistis et al., 2015), several studies have reported that larger, population-specific reference panels, such as the Haplotype Reference Consortium (HRC—consisting of 64,976 haplotypes from 32,611 individuals of predominantly European ancestry (McCarthy et al., 2016), may enhance imputation quality, especially for rare and low-frequency variants (Kim et al., 2015; Li et al., 2011; Martin et al., 2014; Tantoso et al., 2014; Zheng et al., 2015) and improve the identification of novel loci not detectable with the exome array alone and/or find causal variants using fine mapping.
The Beaver Dam Eye Study (BDES) is a population-based ocular cohort study of primarily European–American adults from Beaver Dam, Wisconsin. Exome variant and gene-level quantitative trait association analyses have been done in this population investigating variants associated with mean spherical equivalent revealing several significantly associated genes (Chen et al., 2016). In the current study, BDES participants were genotyped on the Illumina exome array and densely imputed using the HRC panel to evaluate three ocular phenotypes: spherical equivalent, axial length, and corneal curvature.
2 MATERIALS AND METHODS
2.1 Editorial policy and ethical considerations
Data were collected according to the tenets of the Declaration of Helsinki, and all protocols were prospectively approved by the institutional review board at the University of Wisconsin at Madison. All study participants provided informed consent.
2.2 The Beaver Dam Eye Study (BDES)
The BDES has been described previously (Klein, Klein, & Lee, 1996; Klein, Klein, Lee, Cruickshanks, & Chappell, 2001; Klein, Klein, Lee, Cruickshanks, & Gangnon, 2006; Klein, Klein, Linton, & De Mets, 1991; Klein, Lee, Gangnon, & Klein, 2013; Linton, Klein, & Klein, 1991). Briefly, the baseline examination of the BDES included a personal history questionnaire and a complete ocular examination. The eye exam included a standardized evaluation of refraction using the Humphrey 530 refractor (Humphrey Instruments Inc., San Leandro, CA) (Linton et al., 1991). A standard formula was used to calculate the mean spherical equivalent, in diopters (D) for each eye (sphere +0.5 × cylinder). Nuclear sclerosis was measured as the sum of the nuclear lens opacity score of both eyes at baseline (as described in Supporting Information). During the fourth visit, ocular biometry measurements, including axial length and corneal curvature, were obtained using partial coherence laser interferometry (IOLMaster, Carl Zeiss Meditec, Jena, Germany). Age, sex, height, and years of education were obtained at all visits.
2.3 Trait definitions
Axial length, corneal curvature, and spherical equivalent were each analyzed independently. Analyses of axial length and corneal curvature were conducted using measurements of the right eye at fourth visit, followed by analyses using measures of the left eye at fourth visit to ensure consistency of results (Klein et al., 2009). Spherical equivalent was measured as the average spherical equivalent between the right and left eyes at baseline (Chen et al., 2016). Nuclear sclerosis was measured as the sum of the nuclear lens opacity score of both eyes at baseline. We included age and sex for the model in the three traits. Additionally, we included education in the model to analyze axial length; height for corneal curvature and education, and nuclear sclerosis for spherical equivalent (Supporting Information).
2.4 Quality control for samples for axial length, corneal curvature, and spherical equivalent
One individual from each first- or second-degree relative pair was removed following imputation on 1,871 individuals (n = 117), as were all individuals with missing values of the relevant phenotype and covariates. Individuals with differences in axial length or corneal curvature >3 SDs from the mean between the two eyes were excluded from each respective dataset. Similarly, individuals with differences in spherical equivalent >±4 D between the left and right eyes were removed from the third dataset. Final sample sizes for axial length, corneal curvature, and spherical equivalent were 874, 883, and 1,552 individuals, respectively.
2.5 Quality control for genotyping and imputation
A nested subset of 2,032 individuals from the BDES was genotyped on the Illumina Infinium HumanExome-12 v1.1 BeadChip (Illumina, Inc., San Diego, CA) exome array. Individuals were originally selected based on the tail ends of the distribution for the quantitative traits of intraocular pressure and spherical equivalent. The distribution of both traits was normal following selection. Genotyping was conducted at the Genetic Resource Core Facility, Johns Hopkins Institute of Genetic Medicine. Genotype calling was performed using Illumina's GenTrain clustering algorithm in GenomeStudio.
Among the 1,908 individuals who were successfully genotyped (sample call rate > 98%), some individuals were removed for quality control: unresolved sex inconsistencies (n = 15), Mendelian errors (n = 2), and unexpected duplicates (n = 8). Principal component analysis using SMARTPCA in EIGENSTRAT version 4.2 (Supporting Information, Figure S1) (Price et al., 2006) identified 12 individuals of non-European ancestry who were excluded from further analysis.
Variants were excluded if they were nonautosomal (n = 5,465), call rates < 98% (n = 6,114), out of Hardy–Weinberg Equilibrium (P < 1.0 × 10−6) (Wigginton, Cutler, & Abecasis, 2005) (n = 68), monomorphic (n = 135,499), indels (n = 61), or duplicates (n = 318). The remaining 1,871 individuals and 95,376 variants were imputed to the HRC reference panel (n = 32,611) using SHAPEIT version 2 and Minimac version 3 on the publicly available Michigan Imputation Server (Das et al., 2016; Delaneau et al., 2012, 2013; McCarthy et al., 2016). A total of 1,024,985 predominantly exonic variants were used for the association analyses after applying an imputation R2 threshold ≥ 0.6 (McCarthy et al., 2016; Pistis et al., 2015). Filtering and formatting of the datasets were performed with PLINK and VCFtools (Danecek et al., 2011; Purcell et al., 2007). Imputed dosages were used to account for imputation uncertainty. DosageConverter version 1 (DosageConverter, University of Michigan, Ann Arbor, MI) was used to convert the variant call files from the Michigan Imputation Server to dosage files.
2.6 Single-variant analysis
We conducted a single-variant analysis under an additive model using linear regression in Mach2qtl (Li, Willer, Ding, Scheet, & Abecasis, 2010; Pei, Zhang, Li, & Deng, 2010).
Single-variant analyses were performed on rare (0.0057 ≤ MAF < 0.01), low-frequency (0.01 ≤ MAF ≤ 0.05), and common (MAF > 0.05) variants for axial length and corneal curvature. Rare variants were defined as 0.0032 ≤ MAF < 0.01 for spherical equivalent. Variants with a MAF ˂ 0.0057 (axial length and corneal curvature) or 0.0032 (spherical equivalent) thresholds (minor allele count < 10) were not considered (Chen et al., 2014, 2016; Soderholm et al., 2016). A Bonferroni-corrected significance threshold of P < 1.09 × 10−6 for axial length and corneal curvature, and P < 9.25 × 10−7 for spherical equivalent, were used based on the number of independent markers in each dataset (linkage disequilibrium r2 < 0.20). Variants with MAF > 0.01 and effect sizes ≥ 1.5 for axial length, ≥ 0.4 for corneal curvature, or ≥ 3.0 for spherical equivalent could be detected with sufficient power (80%) in these single-variant analyses (Gauderman & Morrison, 2006). Significantly associated variants and suggestively associated variants (P < 1.0 × 10−4) are reported.
2.7 Gene-based analysis
We conducted an optimized sequence kernel association test (SKAT-O) using the default weight 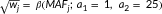 to upweight rare variants presumed to be causal (Lee, Miropolsky, & Wu, 2014; Lee, Wu, & Lin, 2012). The a1 and a2 parameters represent the
to upweight rare variants presumed to be causal (Lee, Miropolsky, & Wu, 2014; Lee, Wu, & Lin, 2012). The a1 and a2 parameters represent the  distribution shape parameters α and β, respectively. The gene-based SKAT-O test was conducted in the Efficient and Parallelizable Association Container Toolbox (University of Michigan) program, using genotype dosages to account for imputation uncertainty. Gene groups were defined using variant annotations in ANNOVAR under GRCh37/hg19 (Wang, Li, & Hakonarson, 2010). Genes with more than one rare or low-frequency variant (MAF ≤ 0.05) were included in the analysis. Genes with a cumulative MAF (CMAF, the sum of MAF values of all variants within the gene) > 0.01 are reported as reliable findings. A total of 13,854 autosomal genes were tested for their association with axial length and corneal curvature, and 13,868 genes were tested for their association with spherical equivalent. This translated to a Bonferroni-corrected significance threshold of P < 3.61 × 10−6. Significantly associated genes and suggestively associated genes (P < 1.0 × 10−4) are reported.
distribution shape parameters α and β, respectively. The gene-based SKAT-O test was conducted in the Efficient and Parallelizable Association Container Toolbox (University of Michigan) program, using genotype dosages to account for imputation uncertainty. Gene groups were defined using variant annotations in ANNOVAR under GRCh37/hg19 (Wang, Li, & Hakonarson, 2010). Genes with more than one rare or low-frequency variant (MAF ≤ 0.05) were included in the analysis. Genes with a cumulative MAF (CMAF, the sum of MAF values of all variants within the gene) > 0.01 are reported as reliable findings. A total of 13,854 autosomal genes were tested for their association with axial length and corneal curvature, and 13,868 genes were tested for their association with spherical equivalent. This translated to a Bonferroni-corrected significance threshold of P < 3.61 × 10−6. Significantly associated genes and suggestively associated genes (P < 1.0 × 10−4) are reported.
2.8 Association of known genome-wide association study (GWAS) loci with axial length, corneal curvature, and spherical equivalent
We identified previously published genome-wide association study (GWAS) loci for (1) axial length, (2) corneal curvature, and (3) spherical equivalent. We assessed whether (1) the exact variant, or (2) variants in strong linkage disequilibrium (r2 ≥ 0.7) with these previously published GWAS loci were significantly associated with one or more of the three phenotypes (axial length, corneal curvature, spherical equivalent) analyzed in this study at P < 0.01. Ensembl release 85 (Ensembl, Wellcome Trust Genome Campus, Hinxton, UK) was used to identify variants in linkage disequilibrium (LD) with previously published GWAS variants. Results from the single-variant analyses conducted on the imputed data (n = 874, 883, and 1,552 for axial length, corneal curvature, and spherical equivalent, respectively) were used for analysis of known variants.
2.9 Annotations
SeattleSeq Annotation Server 138 version 9.01 (University of Washington, Seattle, WA) was used to annotate reported genetic variants from the single-variant analyses under GRCh37/hg19. Chromosomal locations and amino acid alterations of variants were obtained using dbSNP build 147 (dbSNP Short Genetic Variations, National Center for Biotechnology Information, Bethesda, MD).
3 RESULTS
3.1 Study participants
The number of individuals analyzed was 874 for axial length, 883 for corneal curvature, and 1,552 for spherical equivalent. Characteristics of the study participants for each phenotype are presented in Table 1 including information about age, gender, height, education, and particular metrics for each trait.
| Characteristica | |
|---|---|
| BDES participants contributing to the axial length analyses (N = 874) | |
| Age (years), mean ± SD (range) | 70.1 ± 8.2 (58–94) |
| Female gender, N (%) | 526 (60.2) |
| Education (years), mean ± SD (range) | 13.2 ± 2.8 (5–25) |
| Height (cm), mean ± SD (range) | 167.4 ± 9.1 (143.0–208.3) |
| Axial length of right eye (mm), mean ± SD (range) | 23.8 ± 1.1 (20.5–28.4) |
| Axial length of left eye (mm), mean ± SD (range) | 23.8 ± 1.2 (20.7–28.2) |
| BDES participants contributing to the corneal curvature analyses (N = 883) | |
| Age (years), mean ± SD (range) | 70.1 ± 8.1 (58–94) |
| Female gender, N (%) | 536 (60.7) |
| Height (cm), mean ± SD (range) | 167.4 ± 9.1 (143.0–208.3) |
| Corneal curvature of right eye (mm), mean ± SD (range) | 7.7 ± 0.3 (7.0–8.6) |
| Corneal curvature of left eye (mm), mean ± SD (range) | 7.7 ± 0.3 (6.9–8.7) |
| BDES participants contributing to the spherical equivalent analyses (N = 1,552) | |
| Age (years), mean ± SD (range) | 59.3 ± 10.5 (43–85) |
| Female gender, N (%) | 896 (57.7) |
| Education (years), mean ± SD (range) | 12.7 ± 3.0 (2–26) |
| Nuclear sclerosis of right eye, mean ± SD (range) | 2.3 ± 0.9 (1.0–5.0) |
| Nuclear sclerosis of left eye, mean ± SD (range) | 2.3 ± 0.9 (1.0–5.0) |
| Nuclear sclerosis, mean ± SD (range) | 4.7 ± 1.6 (2.0–9.0) |
| Spherical equivalent of right eye (D), mean ± SD (range) | −0.3 ± 3.0 (−13.5 to 11.0) |
| Spherical equivalent of left eye (D), mean ± SD (range) | −0.2 ± 3.0 (−13.5 to 11.0) |
| Spherical equivalent (d), mean ± SD (range) | −0.2 ± 2.9 (−13.0 to 10.3) |
- a All measurements were taken from fourth visit except height, which was measured at baseline.
3.2 Single-variant and gene-based analyses
No significant associations of single variants or genes (CMAF > 0.01) with axial length, corneal curvature, or spherical equivalent were identified. Suggestively associated independent variants and gene-based loci are reported in Supporting Information, Tables S1–S4. Results of the right and left eyes were similar for both axial length and corneal curvature, so only the results of the right eye are reported for suggestively associated variants and genes.
3.3 Association with known GWAS genetic loci
We identified 25 axial lengths (11 loci), seven corneal curvature (three loci), and 65 spherical equivalents (50 loci) previously published variants. Several previously published variants associated with corneal curvature (n = 3) and spherical equivalent (n = 2) were also associated in this study at P ≤ 0.01 (Table 2). Imputation increased the coverage of the regions harboring the replicated variants. There were 12 genotyped variants in the chr4q12 region (54.8 MB–55.4 MB) and the number of variants increased to 211 after imputation. Similarly, the number of variants in the chr15q14 region went from 10 genotyped to 32 genotyped variants and imputed. The coverage of the intergenic region was also improved (Supporting Information, Figure S3). The imputed variants were in linkage disequilibrium with a genotyped variant from the array.
| Locusa | GWAS Varianta,b | GWAS Sample size(ref) | Effect allele | GWAS Effect β (CI) | GWAS P value | GWAS Effect allele freqc | BDES Effect β (CI)d | BDES P valuee | BDES Effect allele freq |
|---|---|---|---|---|---|---|---|---|---|
| Corneal curvature score (n = 883) | |||||||||
| 4q12 | rs2114039 g.55092626T > C | 10,008f | C | −0.13 (−0.19, −0.06) | 1.33 × 10−9 | 0.27–0.30 | −0.03 (−0.06, −0.01) | 0.01 | 0.28 |
| 4q12 | rs17084051 g.55087581C > A | 10,008f | A | −0.15 (−0.22, −0.09) | 2.23 × 10−8 | 0.20–0.26 | −0.05 (−0.01, −0.08) | 0.01 | 0.27 |
| 4q12 | rs17084051 g.55087581C > A | 9,383g | A | −0.13 (−0.16, −0.09) | 4.50 × 10−14 | 0.20–0.26 | −0.05 (−0.01, −0.08) | 0.01 | 0.27 |
| 4q12 | rs1800813 g.55094467G > A | 9,913h | G | 0.10 (0.06, 0.14) | 7.50 × 10−9 | 0.77 | 0.05 (0.01, 0.08) | 0.01 | 0.73 |
| Spherical equivalent (n = 1,552) | |||||||||
| 15q14 | rs634990 g.35006073T > C | 15,608i | C | −0.23 (−0.23, −0.23) | 2.21 × 10−14 | 0.47 | −0.27 (−0.45, −0.09) | 3.79 × 10−3 | 0.47 |
| 15q14 | rs685352 g.35008335A > G | 15,608i | G | −0.21 (−0.21, −0.21) | 4.19 × 10−12 | 0.44 | −0.29 (−0.10, −0.49) | 3.12 × 10−3 | 0.47 |
| 15q14 | rs685352 g.35008335A > G | 26,615j | G | −0.08 (−0.01, −0.06) | 2.09 × 10−10 | 0.46 | −0.29 (−0.10, −0.49) | 3.12 × 10−3 | 0.47 |
- GWAS, genome-wide association study; CI, confidence interval; OR, odds ratio; HR, hazard ratio; freq, frequency; BDES, Beaver Dam Eye Study.
- a Reported in GRCh37 (hg19).
- b Variants listed in bold were genotyped on the exome array in the original BDES group of 1,871 individuals. The remaining variants were imputed. The variants in bold represent the independent loci found as associated for each phenotype, while the imputed variants at each locus are in high LD (r2 ≥ 0.7) with these variants.
- c GWAS allele frequency in cases and controls.
- d Single-variant associations of corneal curvature adjusted for age and sex measured at fourth visit, and baseline height; single-variant associations for spherical equivalent adjusted for baseline age, sex, education, and nuclear sclerosis.
- e Significant P value threshold ≤0.01.
- f Han et al., 2011.
- g Guggenheim, McMahon et al., 2013.
- h Chen et al., 2014.
- i Solouki et al., 2010.
- j Verhoeven et al., 2012.
rs2114039 (g.55092626T > C), which is in an intergenic region on chromosome 4q12 near the platelet-derived growth factor receptor α (PDGFRA) gene (MIM 473190), was genotyped on the exome array and was significantly associated with corneal curvature in a previous GWAS and replicated in this study with the same direction of effect at P ≤ 0.01 (Han et al., 2011) (Table 2). Similarly, two imputed variants (rs17084051, g.55087581C > A and rs1800813, g.55094467G > A; imputation R2 = 0.72) in moderate LD with rs2114039 (r2 = 0.66–0.70) and previously associated with corneal curvature were also associated with corneal curvature in this study with the same direction of the effect (Table 2) (Guggenheim et al., 2013; Han et al., 2011). The effect of those variants in our study is lower compared to the GWAS effect.
Two variants in high LD (r2 = 0.85) on chromosome 15q14 near the gap junction protein, delta-2 (GJD2) gene (MIM 607058) that were associated with spherical equivalent in previously published studies were also associated with spherical equivalent at P < 0.01 in this study (Table 2) (Solouki et al., 2010; Verhoeven et al., 2012). Rs634990 (g.35006073T > C) was genotyped on the exome array and rs685352 and g.35008335A > G was imputed. Both showed a consistent direction of effect with the previously published study (Table 2). We were unable to find any association with variants previously associated with axial length in this study (Supporting Information, Table S5).
4 DISCUSSION
In the current study, we performed single-variant and gene-based analyses of genotyped and imputed exome variants to evaluate associations with spherical equivalent, axial length, and corneal curvature in a European–American population. We did not identify new loci associated with these traits. However, we replicated previous associations with two of the three traits.
We replicated a locus associated with corneal curvature located on chromosome 4q12 near the PDGFRA gene. The three variants showed a consistent direction of effect with the previous published studies (Guggenheim et al., 2013; Han et al., 2011). However, the effect was attenuated in our study. We consider that the differences in the effect size could be attributed to the demographic characteristic of the compared populations. The reference studies include European young individuals (mean age: 15.5 years) and Asians in a wide range of age (10–47 years) and the BDES study is comprised of older individuals. Also, the distribution of corneal curvature is different between the studies with similar means (∼7.7 mm) for both traits but with a higher variance in our cohort.
We identified two variants previously associated with spherical equivalent in our study. All variants were located within the same genetic region (Chen et al., 2016; Kiefer et al., 2013; Solouki et al., 2010). One of them (rs634990) was genotyped on the exome array and the other (rs685352) was imputed. Both are on chromosome 15q14, 39 kb upstream from the 3′ end of the GJD2 gene. Mutations in the GJD2 gene have been repeatedly associated with refractive error and degenerative myopia (“GeneCards Human Genome Database,” 2017; Solouki et al., 2010). The imputed variant was initially significantly associated in a Dutch population-based study with replication in four independent cohorts with 15,608 individuals in the meta-analysis of all cohorts (Solouki et al., 2010). The effect in the meta-analysis was lower than in the current study, however, it showed some variation in the European cohorts included (−0.05 to 0.33) depending on the cohort. This range is consistent with the value obtained in this study adding reliability to our findings.
The same variant was also detected in a meta-analysis (n = 49,363) including 26,615 Caucasians (Verhoeven et al., 2012). Although the minor allele, MAF, adjusted covariates, and directions of effect presented for this variant are consistent between these two published studies, the magnitudes of effect are less consistent (Solouki et al., 2010; Verhoeven et al., 2012). In our case, we observed a larger effect compared to the meta-analysis presented by Verhoeven et al. However, that analysis included multiple cohorts across Europe and Asians with different characteristic regarding age, height, and sex with internal variability in the findings: the effect size of the discovery population was −0.20, −0.08 in the (larger) replication, and −0.11 in the Asian population. The studies would likely be more comparable if they were homogeneous cohorts, such as the BDES.
We did not replicate any previously associated variants with axial length in our study (Cheng et al., 2013; Fan et al., 2012). With a MAF of 0.3, the sample size, and mean axial length from our axial length sample, we calculated that we have 80% power to detect significant associations (at P < 0.01) only if effect sizes were ˃0.20. This effect size is larger than those reported in the current study; thus, we did not have sufficient power to detect associations with variants previously reported with axial length.
In summary, we replicated variants within previously published genetic regions after increasing the coverage with imputed variants. The replicated variants were in high LD with associated variants genotyped on the exome array. This finding confirms these loci as etiologically relevant for the associated ocular traits. Further analysis of the replicated loci would potentially help to understand relevant biological mechanisms underlying these phenotypes.
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Eye Institute of the National Institutes of Health under grant award numbers R01EY021531, U10006594 and 1T32EI022303; and by the Research to Prevent Blindness Unrestricted Grant to the Department of Ophthalmology and Visual Sciences, University of Wisconsin. We acknowledge the editorial contributions of Nicole Thornton. The authors are grateful to the study participants and thank the staff and investigators of the Beaver Dam Eye Study.
CONFLICTS OF INTEREST
The authors declare no conflict of interest.