Characterization of spliceogenic variants located in regions linked to high levels of alternative splicing: BRCA2 c.7976+5G > T as a case study
Gemma Montalban and Eugenia Fraile-Bethencourt contributed equally to this work.
Communicating by Marc S. Greenblatt
Abstract
Many BRCA1 and BRCA2 (BRCA1/2) genetic variants have been studied at mRNA level and linked to hereditary breast and ovarian cancer due to splicing alteration. In silico tools are reliable when assessing variants located in consensus splice sites, but we may identify variants in complex genomic contexts for which bioinformatics is not precise enough. In this study, we characterize BRCA2 c.7976 + 5G > T variant located in intron 17 which has an atypical donor site (GC). This variant was identified in three unrelated Spanish families and we have detected exon 17 skipping as the predominant transcript occurring in carriers. We have also detected several isoforms (Δ16-18, Δ17,18, Δ18, and ▼17q224) at different expression levels among carriers and controls. This study remarks the challenge of interpreting genetic variants when multiple alternative isoforms are present, and that caution must be taken when using in silico tools to identify potential spliceogenic variants located in GC-AG introns.
BRCA1 (MIM# 113705) and BRCA2 (MIM# 600185; BRCA1/2) genes are associated to hereditary breast and ovarian cancer syndrome (HBOC). Pathogenic variants in BRCA1/2 result in an increased cumulative breast cancer risk to age 80 that ranges from 61% to 79%, and in an increased risk for ovarian cancer that ranges from 11% to 53% (Kuchenbaecker et al., 2017). Genetic variants in disease-responsible genes that disrupt the splicing code have a key role in human hereditary disorders and cancer. A recent worldwide study describing the mutational spectrum of BRCA1/2 genes in HBOC families identified that 10.1% of BRCA1 and 7.6% of BRCA2 pathogenic variants result in aberrant mRNA splicing (Rebbeck et al., 2018). Splicing mutations were traditionally considered those that affect consensus splice sites (intronic nucleotides +1 and +2 of the donor GT, and −1 and −2 of the acceptor AG sites), but other intronic and exonic nucleotides outside these regions have also been found to be highly conserved and critical for splice site selection (Cartegni, Chew, & Krainer, 2002; Manning & Cooper, 2017). More than 99% of human introns are flanked by GT-AG splice site dinucleotides and are spliced by the so-called major U2-type spliceosome. An exception to this rule are the U2-type GC-AG introns, such as BRCA2 intron 17 (see Supporting Information Figure S1), comprising about 0.9% of all human splice sites (reviewed by Parada, Munita, Cerda, & Gysling, 2014; Sibley, Blazquez, & Ule, 2016). GC-AG introns possess weak donor sites that are compensated with strong consensus in the surrounding nucleotides of the donor, and are usually linked to alternatively spliced exons (Churbanov, Winters-Hilt, Koonin, & Rogozin, 2008; Kralovicova et al., 2011; Thanaraj & Clark, 2001). Given the complexity of these introns, we aimed to characterize the splicing impact of BRCA2 c.7976 + 5G > T variant, located at position +5 from the atypical donor site of intron 17. To our knowledge, this variant is not present in the gene-specific databases LOVD (www.lovd.nl), BRCA Exchange (brcaexchange.org), and BIC (https://research.nhgri.nih.gov/bic), ascertained by February 2018. This variant is reported twice in ClinVar database and categorized as conflicting (https://www.ncbi.nlm.nih.gov/clinvar/), it is reported as a variant of unknown significance (VUS) in BRCA Share (www.umd.be), and it is classified as likely benign (Class-2) in a previous work published by Garibay et al., 2014.
We identified this variant in three unrelated HBOC Spanish families and initiated this collaborative study to exhaustively re-evaluate the variant across three laboratories: Hospital Universitari Vall d'Hebron (HUVH), Hospital Clínico San Carlos (HCSC), and Instituto de Biología y Genética Molecular (IBGM). Probands underwent genetic counselling and written informed consent was obtained in all cases. Family information and pedigrees are described in Supporting Information Figure S2.
BRCA2 c.7976 + 5G > T was first assessed in silico using Human Splicing Finder (HSF) and MaxEntScan (MES) (https://www.umd.be/HSF3/), which predicted a reduction of the native donor splice site (11.66% and 74.19%, respectively). However, when using the splicing module of Alamut software v2.10 (Interactive Biosoftware) only SSF-like computed a score, predicting a 12.8% reduction of the donor site (Supporting Information Table S1). Breast Cancer Genes Prior Probabilities website (https://priors.hci.utah.edu/PRIORS/index.php) also predicted a high probability of pathogenicity (0.97) due to splice site donor damage. In vitro characterization was performed with patient RNA and a minigene system (see Supporting Information Methods and Supporting Information Table S2 for detailed protocols used in each laboratory). HUVH samples were analyzed by RT-PCR using primers located in exons 15 and 19. We detected five transcripts corresponding to the reference full-length (FL; 906 bp), Δ18 (551 bp), Δ17 (735 bp), Δ17,18 (380 bp), and Δ16-18 (192 bp; Supporting Information Figure S3). All transcripts were detected in carrier and control samples (n = 10), with the exception of Δ17, which was only present in the carrier. Capillary electrophoresis of fluorescent amplicons (CE) ruled out any expression of Δ17 in control samples, indicating that only the variant allele generates this transcript (Figure 1a). HCSC samples were analyzed using primers located in exons 16 and 19. In variant carrier, RT-PCR assays (Figure 1b) showed transcripts corresponding to FL, Δ17, Δ17,18, and an additional peak of ≈900 nt that we tentatively annotated as ▼17q224 (Supporting Information Figure S4). In controls (n = 34), FL was detected in all samples, Δ17 was absent in all samples, and isoforms Δ17,18, Δ18 and the putative ▼17q224 were detected in 11, 3, and 6 samples, respectively. IBGM carrier was analyzed with primers located in exons 16 and 19 and RT-PCRs showed FL and Δ17 transcripts, without evidence of any other aberrant transcripts or isoforms (Figure 1c). In summary, we identified three frameshift isoforms Δ17,18 (p.Ala2603Phefs*43), Δ18 (p.Tyr2660Phefs*43), and ▼17q224 (p.Arg2659Argfs*3) occurring in carrier and control samples; one in-frame isoform Δ16-18 (p.Leu2540_Lys2777del) detected in carrier and control samples; and one in-frame isoform Δ17 (p.Ala2603_Arg2659del) detected only in carriers (results are summarized in Supporting Information Table S3). All transcripts were confirmed by Sanger sequencing with the exception of Δ16-18 and ▼17q224, which were imputed based on the length of the product observed.
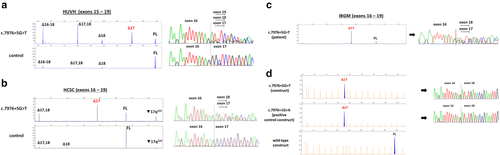
Semi-quantitative CE analysis was performed in HUVH and HCSC samples (IBGM patient sample was not available) and showed that Δ17 represents a substantial contribution to the total splicing fraction (SF) in HUVH carrier and HCSC carrier (average SF 34.3% and 48.3%, respectively), whereas in controls was not detected (Figure 2a). Regarding alternative transcripts, notable differences were observed in isoform Δ17,18 levels between HUVH and HCSC carriers (SF 26% and 9.6%, respectively), whereas HUVH and HCSC controls had similar levels (SF 3.3% and 3.9%, respectively). Isoform Δ18 is a minor event detected only in HUVH carrier (SF 7.2%) and in HUVH/HCSC controls (SF 4.2% and 1.4%, respectively); isoform Δ16-18 was only detected in HUVH due to primer location and displayed no notable differences between carrier and controls (SF 13.1% and 11.1%, respectively); and putative isoform ▼17q224 was only detected in HCSC samples, although not in all RT-PCR assays, and showed higher levels in HCSC carrier (SF 17.1%) compared to controls (1.2%; Figure 2a). Two previous studies detected Δ17,18 and Δ18 in control lymphoblastoid cell lines (LCLs) and normal breast tissue, with Δ17,18 being more abundant (Davy et al., 2017; Fackenthal et al., 2016). Normalized CE data from full-length transcript (FL) showed a two-fold reduction in carriers compared to controls, suggesting that the variant allele is not producing FL (Figure 2b). To test this, since allele-specific assays could not be performed due to the lack of heterozygous informative loci in patient sample, the mutant allele (c.7976 + 5G > T) was artificially interrogated using a pSAD-derived minigene with BRCA2 exons 14 to 20, constructed and functionally validated as previously described in Fraile-Bethencourt et al., 2017. A wild-type (wt) construct and a variant construct BRCA2 c.7976 + 1G > A were used as negative and positive controls, respectively. Wt construct produced a stable canonical transcript of the expected size and structure, and variant constructs revealed a unique transcript corresponding to exon 17 skipping (Mean ± SEM of relative fluorescence: 1.045 ± 0.111 and 0.955 ± 0.020 for variant and positive control, respectively; Figure 1D).
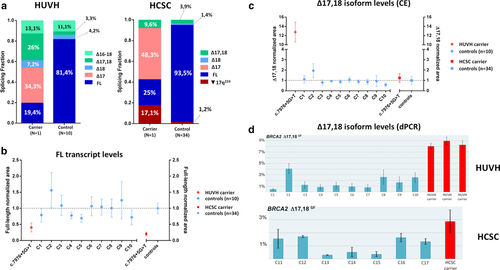
Given that notable differences in isoform Δ17,18 levels were observed among samples (Figure 2a), we aimed to determine whether such differences were due to technical reasons and we used digital PCR as a second approach to measure Δ17,18 levels. Data obtained was consistent with CE data, i.e., the highest value was observed in HUVH carrier (SF ∼ 9% vs. SF ∼ 2% observed in HCSC carrier) (Figures 2c and d). A slight variability was also observed among control samples, with SF levels ranging from ∼0.5% to 4%. Interestingly, a recent study identified a common BRCA2 variant c.7806-14T > C (rs9534262) which influences Δ17,18 levels by modulating exon 17 acceptor site (Garibay et al., 2014). DNA sequence analysis revealed that HUVH patient is homozygous (C/C) at this polymorphic position, whereas HCSC and IBGM are heterozygous (T/C; data not shown). Sanger sequencing of introns surrounding exon 17 (limited to c.7806−37_7806−1 and c.7976 + 1_7976 + 25 regions) in HUVH and HCSC carriers, and whole intron 16 and 17 analysis in IBGM carrier, did not identify additional rare or common variants that could explain differential Δ17,18 isoform expression among carriers. In this regard, patient genotypes were consistent with experimental data in that the highest Δ17,18 levels were detected in the homozygous C/C patient. The influence of c.7806 − 14T > C on Δ17,18 levels was additionally evaluated in HCSC controls (n = 34) by RT-PCR and CE, and the highest levels were again observed in C/C samples (Supporting Information Figure S5). Nevertheless, whether an increase of Δ17,18 is associated with our variant under scrutiny or not, would require the analysis of carriers being c.7806-14T/T to avoid its influence on Δ17,18 levels.
We also re-evaluated HUVH and HCSC RNA samples to explore whether methodological differences between laboratories could influence variability in isoform levels. To do so, carriers and controls were analyzed using primers and protocols from HCSC laboratory (see Supporting Information Table S2). Results obtained showed that major transcriptional events were detected as previously: Δ17 was present in both carriers and absent in controls, and Δ17,18 highest levels were detected in HUVH carrier. Minor isoform Δ18 was again detected in HUVH carrier and absent in HCSC carrier, and ▼17q224 was not detected in either carriers (Supporting Information Figure S6). This data indicates that differences in minor events (Δ18 and ▼17q224) are presumably due to stochastic effects during PCR amplification, but variability in major isoform Δ17,18 is likely to be linked to individual genetic features rather than methodological differences. In this analysis, polymerases and CE conditions did not seem to have an influence in isoform detection, and although new RNA samples could not be obtained to rule out any influence of RNA isolation methods, a collaborative work comparing RNA protocols for characterization of spliceogenic variants across multiple laboratories concluded that RNA extraction methods were indistinguishable (Whiley et al., 2014).
Overall, combined analysis carried out in three different laboratories provides convincing evidence that the major splicing outcome produced by BRCA2 c.7976 + 5G > T variant is exon 17 skipping, which causes an in-frame deletion (r.7806_7976del171) that results in a protein lacking 57 amino acids (aa) (p.Ala2603_Arg2659del). The lost region is part of the α-helical domain (aa 2479 to 2667) of BRCA2 DNA Binding Domain (DBD), which has 30 strictly conserved residues from sea urchin to human (Supporting Information Figure S7). This domain enables BRCA2 binding to single-stranded and double-stranded DNA, and is essential to allow DNA repair by homologous recombination (HR) (Roy, Chun, & Powell, 2011). Functional assays for classification of missense variants located in this region confirmed a reduction of BRCA2 HR activity for Trp2626Cys, Ile2627Phe, Leu2647Pro, Leu2653Pro, and Arg2659Lys variants (Biswas et al., 2011; Farrugia et al., 2008). From these, variants Trp2626Cys, Ile2627Phe, Leu2653Pro, and Arg2659Lys had been previously evaluated by multifactorial analysis and classified as pathogenic (Class 5) (Easton et al., 2007). Other variants causing exon 17 skipping (c.7976G > A, c.7976 + 1G > A and c.7976 + 3_7976 + 4del) have been identified in HBOC patients and reported as deleterious (Brandão, Roozendaal, Tserpelis, García, & Blok, 2011; Fraile-Bethencourt et al., 2017; Hofmann, Horn, Hüttner, Classen, & Scherneck, 2003; Thirthagiri et al., 2008; Wu et al., 2005). Moreover, allele-specific assessment was performed in patient RNA for variant c.7976 + 3_7976 + 4del, and only detected transcript lacking exon 17 (Brandão et al., 2011). Although Δ17 was categorized as a minor alternative splicing event occurring in control LCLs and normal breast tissue, it was not detected in whole blood control samples (Fackenthal et al., 2016). Likewise, we did not identify this transcript in our control group (n = 44).
Our variant G > T is located at position +5 from intron 17 donor splice site, where a G is present in > 80% of human introns (Zhang, 1998). In this particular case, BRCA2 intron 17 has an atypical donor site GC weaker than the GT counterparts because of the +2 substitution, meaning that the rest of nucleotide positions are more conserved (Thanaraj & Clark, 2001) and that variants in any of these nucleotides may have an impact on exon recognition. However, it is important to note that not all in silico approaches used in this study to predict the splicing impact of BRCA2 c.7976 + 5G > T variant were able to detect its potential damage to the native donor site. Other BRCA1/2 variants located at position +5 have been reported to induce splicing alterations, such as BRCA1 c.5406 + 5G > C (exon 22 skipping) and c.5467 + 5G > C (exon 23 skipping), BRCA2 c.316 + 5G > C (exon 3 skipping), and c.8754+5G > A (insertion of 46 nucleotides of intron 21) (Acedo, Hernández-Moro, Curiel-García, Díez-Gómez, & Velasco, 2015; Houdayer et al., 2012; Vreeswijk et al., 2009; Whiley et al., 2011). Spliceogenic variants in +5 positions have also been reported for other disease-responsible genes such as CFTR c.2657+5G > A (exon 14b skipping) (Highsmith et al., 1997) and MLH1 c.116 + 5G > C (retention of 227 intronic nucleotides) (Arnold et al., 2009). These results highlight the need to study potential splicing alterations beyond consensus positions GT-AG, and that special caution must be taken when relying on in silico predictions to detect potential spliceogenic variants located in GC-AG introns.
In summary, our splicing analysis performed in three independent carriers shows that BRCA2 c.7976 + 5G > T alleles produce a major in-frame transcript Δ17 predicted to encode a nonfunctional protein, and that even though variable proportions of additional transcripts (Δ16-18, Δ17,18, Δ18, and ▼17q224) have been detected, none of them are predicted to rescue BRCA2 functionality.
According to ACMG-AMP guidelines (Richards et al., 2015), our variant qualifies for categories PS3 (“Well-established in vitro or in vivo functional studies supportive of a damaging effect on the gene or gene product”), PM2 (“Absent from controls [or at extremely low frequency if recessive] in Exome Sequencing Project, 1000 Genomes Project, or Exome Aggregation Consortium”), and PP4 (“Patient's phenotype or family history is highly specific for a disease with a single genetic etiology”). Combining these criteria, the variant is classified as likely pathogenic.
Similarly, following rigorously ENIGMA (Evidence-based Network for the Interpretation of Germline Mutant Alleles; https://enigmaconsortium.org/) classification criteria, the variant should be classified as likely pathogenic (Class 4): “variant considered extremely likely to alter splicing based on position, and is untested for splicing aberrations using in vitro assays of patient RNA that assesses allele-specific transcript expression, and is predicted bioinformatically to alter use of the native donor/acceptor site, and is not predicted or known to alter production of (naturally occurring) in-frame RNA isoforms that may rescue gene functionality”. However, the variant also meets the ENIGMA requirements to be considered pathogenic (Class 5): “Variant allele tested for mRNA aberrations using in vitro assays of patient RNA that assesses allele-specific transcript expression, and is found to produce only transcript(s) carrying a premature termination codon, or an in-frame deletion disrupting expression of one or more known clinically important residues”, with the exception that our in vitro allele-specific analysis was performed in a minigene instead of patient RNA. Current ENIGMA guidelines do not consider construct-based mRNA assays alone as a sufficiently robust approach to be used as evidence for variant classification. However, in this study, we used a validated minigene (MGBR2_ex14-20) that confers high reproducibility of splicing patterns as previously described in Fraile-Bethencourt et al. (2017). More specifically, authors analyzed variants involving exon 17 (c.7806-9T > G, c.7975A > G, c.7976G > A, and c.7976G > C) using the MGBR2_ex14-20 minigene and compared the splicing patterns with patient RNA results published in previous works, and identified the same splicing results for both approaches in all cases. Furthermore, our study by semi-quantitative methods in patient RNA also supports a pathogenic role for the variant given that carriers generate a predominant aberrant transcript, with any other evidence of transcripts that could rescue protein function. In all, we consider that our results obtained with different methodologies are in agreement and robust enough to support the classification of this variant as pathogenic (Class-5).
The clinical interpretation of interindividual differences in isoform expression levels is challenging and whether they are true features associated with the variant of interest, or just reflect biological or technical variability, cannot be concluded from our study. Genetic variants have the potential to generate complex splicing profiles when located in genomic regions with high levels of alternative splicing, and these profiles can be even more complicated to interpret when common variants that modulate isoform levels are present. A comprehensive characterization is therefore always required in these cases, and it is worth to consider whether the individual genetic makeup may have a role in modulating alternative splicing and cancer risk.
ACKNOWLEDGMENTS
The authors acknowledge CELLEX Foundation for providing research facilities and equipment.
CONFLICT OF INTEREST
The authors declare no conflict of interest.