Targeted resequencing reveals genetic risks in patients with sporadic idiopathic pulmonary fibrosis
Communicated by Garry R. Cutting
Funding information:
Integrated Innovative Team for Major Human Diseases Program of Tongji Medical College, Huazhong University of Science and Technology; Clinical Research Physician Program of Tongji Medical College, Huazhong University of Science and Technology.
Abstract
Idiopathic pulmonary fibrosis (IPF) is a genetic heterogeneous disease with high mortality and poor prognosis. However, a large fraction of genetic cause remains unexplained, especially in sporadic IPF (∼80% IPF). By systemically reviewing related literature and potential pathogenic pathways, 92 potentially IPF-related genes were selected and sequenced in genomic DNAs from 253 sporadic IPF patients and 125 matched health controls using targeted massively parallel next-generation sequencing. The identified risk variants were confirmed by Sanger sequencing. We identified two pathogenic and 10 loss-of-function (LOF) candidate variants, accounting for 4.74% (12 out of 253) of all the IPF cases. In burden tests, rare missense variants in three genes (CSF3R, DSP, and LAMA3) were identified that have a statistically significant relationship with IPF. Four common SNPs (rs3737002, rs2296160, rs1800470, and rs35705950) were observed to be statistically associated with increased risk of IPF. In the cumulative risk model, high risk subjects had 3.47-fold (95%CI: 2.07–5.81, P = 2.34 × 10−6) risk of developing IPF compared with low risk subjects. We drafted a comprehensive map of genetic risks (including both rare and common candidate variants) in patients with IPF, which could provide insights to help in understanding mechanisms, providing genetic diagnosis, and predicting risk for IPF.
1 INTRODUCTION
Idiopathic pulmonary fibrosis (IPF) is a chronic fatal interstitial pulmonary disease characterized by the progressive loss of lung function with diagnosis based on clinical and radiologic or histologic criteria (Raghu et al., 2011). Typically, IPF presents as late-onset pulmonary dysfunction with an average onset age of 60–70 years and a median survival of 2–3 years after the initial diagnosis (Raghu et al., 2011). Because the pathogenesis is poorly understood, there are no curative treatments except for lung transplantation (Raghu et al., 2011).
In recent years, there has been growing evidence that genetic factors play an important role in both sporadic and familial IPF cases (Fernandez et al., 2012; Garcia-Sancho et al., 2011). Recent independent studies have shown that up to 20% of IPF patients have a family history and can present earlier, indicating that both the frequency of familial pulmonary fibrosis (FPF) and the genetic risk of sporadic IPF could be underestimated (Fernandez et al., 2012; Garcia-Sancho et al., 2011). Previous investigation of genetic data from FPF cases and sporadic patients have led to the identification of rare pathogenic variants in multiple genes, such as surfactant-associated genes (surfactant protein C, SFTPC; surfactant protein A2, SFTPA2; and ATP-binding cassette member A3, ABCA3) (Lawson et al., 2004; Wang et al., 2009) and telomerase-related genes (telomerase reverse transcriptase, TERT; telomerase RNA component, TERC; regulator of telomere elongation helicase 1, RTEL1) (de Leon et al., 2010). In addition, two large genome-wide association studies (GWASs) conducted in patients with sporadic and familiar IPF not only confirmed known associations with TERC, TERT, and mucin 5B gene (MUC5B), but also found novel variants associated with IPF susceptibility, including variants within toll interacting protein (TOLLIP) and signal peptide peptidase like 2C (SPPL2C) (Fingerlin et al., 2013; Noth et al., 2013). Additionally, a common polymorphism (rs35705950) in the promoter of MUC5B is significantly more prevalent in individuals with both sporadic and familial IPF (Peljto et al., 2013; Zhu et al., 2015). Importantly, pulmonary fibrosis can occur in some rare genetic disorders such as dyskeratosis congenita, Hermansky-Pudlak syndrome (HPS), and tuberous sclerosis, indicating a shared genetic pathogenesis (Hisata et al., 2013; Islam & Roach, 2015; Vicary, Vergne, Santiago-Cornier, Young, & Roman, 2016). Current data suggest that at least one-third of the sporadic and familial IPF can be explained by common genetic variants identified in large GWASs, some of the variants differ in different populations, some associated with disease prognosis or response to treatment (Fingerlin et al., 2013).
Since no previous studies investigated all of the above candidate genes comprehensively in Chinese IPF patients, we used high-throughput targeted-resequencing to sequence 92 potentially IPF-related genes in 253 Chinese patients with IPF and 125 matched controls. We report here the spectrum of variants in these genes and identify novel rare variants and common SNPs that may be potentially associated with IPF.
2 MATERIALS AND METHODS
2.1 Study population
In this study, a total of 253 IPF patients and 125 matched controls were enrolled. All subjects were unrelated, and of Han origin from seven different provinces in mainland China. Criteria for selection of controls are: (1) gender and age-matched, (2) unrelated individuals of Han ancestry, and (3) exclude from pulmonary fibrosis and genetic disease. The diagnostic criteria of IPF cases was based on the ATS/ERS/JRS/ALAT guidelines published in 2011, which include the exclusion of other known causes of interstitial lung disease, the presence of a usual interstitial pneumonia pattern on high-resolution computed tomography (HRCT) in patients not subjected to surgical lung biopsy, specific combinations of HRCT and surgical lung biopsy findings in patients subjected to surgical lung biopsy, and abnormalities of lung function tests (Raghu et al., 2011). At least two experts in pulmonary disease and two radiologists independently reviewed each patient's clinical and biopsy findings and HRCT scans. For each participant, medical history, family history, and other basic information and peripheral blood were collected after informed consent was obtained. Ethical approval for this study was obtained from the Institutional Review Board of Tongji Hospital.
2.2 Gene selection and primer design
We based the selection of potentially IPF-related genes on literature (up to January 2016) and several online databases (OMIM: https://www.ncbi.nlm.nih.gov/omim/; GeneCards: https://www.genecards.org/; HGMD: https://www.hgmd.cf.ac.uk/ac/-index.php; GEO: https://www.ncbi.nlm.nih.gov/geoprofiles/) and our previous IPF data. The target panel included genes accounted for FPF, genes from GWASs and animal experiments, genes for rare genetic syndromes such as dyskeratosis congenita, HPS, and tuberous sclerosis that may be associated with pulmonary fibrosis. This list of 92 genes was submitted to Ion AmpliSeq Designer software (Version 4.24) for primer design (Supp. Table S1). Then primers covering all coding regions and at least 5 bp flanking regulatory regions were synthesized and pooled into two multiplex reactions (Supp. Table S2).
2.3 Library preparation and next-generation sequencing
Genomic DNA (gDNA) was extracted from peripheral blood samples by a Blood DNA kit (TIANGEN BIOTECH, Beijing, China) and was diluted to 5 ng/μl. Then gDNA was amplified and libraries were constructed using the Ion AmpliSeq™ Library Kit 2.0 and customized multiplex PCR primer pools (Life Technologies, Carlsbad, California, USA) on the Ion Torrent platform (Thermo Fisher, San Jose, California, USA) as we previously described (Li et al., 2017). Briefly, after purification, libraries were quantitated using a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, California, USA) and pooled at equal ratios for emulsion PCR on an Ion OneTouch System. Then, templated Ion Sphere particles were enriched by using the Ion OneTouch ES (Life Technologies, Carlsbad, California, USA). The template-positive Ion Sphere particles were loaded for sequencing on the Ion Torrent Proton (Life Technologies, Carlsbad, California, USA).
2.4 Bioinformatics analysis
The process of bioinformatics analysis was shown in the diagram (Supp. Figure S1). Sequencing raw data were first processed with the Ion Torrent Suite v5.0.4 to align to the human genome reference (hg19/GRCh37), to call variants and to analyze coverage. Then, detailed annotation of all variants was processed using Ion reporter v5.0 and Annovar software (2017 July) to obtain information including ExonicFunc, AAChange, minor allele frequency (MAF) in 1000Genome, MAF in the Exome Aggregation Consortium (ExAC) database, MAF in the Exome Sequencing Project (ESP), SIFT and PolyPhen-2 score and prediction, SNP entries and InterVar prediction (Wang, Li, & Hakonarson, 2010). Finally, the degree of conservation across multiple species of the nonsynonymous variants was estimated using GERP++ scores.
The DNA mutation numbering system is based on cDNA sequence, and +1 means the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1.
In filtering, firstly variants with a read depth <20 or with an imbalanced reference/variant allele read depth >3:1 were considered false calls and were removed. Then, we divided the variants into loss-of-function (LOF) variants (frameshift, nonsense, splicing-site, initiation codon break), rare nonsynonymous coding variants, and common variants according to variant type and MAF. Rare nonsynonymous coding variants (MAF ˂ 0.01) were picked out and evaluated by burden tests. Common variants with a MAF ≥ 0.01 in the 1000Genomes Project database, the UCSC common SNP database, the ExAC, or the ESP database were defined as common SNP and were picked out for association analysis. Finally, variants predicted as benign and likely benign by InterVar were removed. Potential functional LOF variants were recorded at a MAF no more than 0.1% among the sequencing population, the ones observed in control groups were filtered out.
We divided the IPF-associated variants into pathogenic, likely pathogenic and LOF (frameshift, nonsense, and splice site) variants according to the ACMG guideline (Richards et al., 2015).
2.5 Sanger sequencing
PCR primers were designed for all LOF variants, likely pathogenic variants, and the MUC5B rs35705950 SNP reported to be associated with IPF were designed using Primer Premier 5.0 software and confirmed to have unique genomic product of sizes between 300–800 bp by UCSC in-silico PCR (https://genome.ucsc.edu/cgi-bin/hgPcr). PCR amplification was optimized in accordance with the manual for Taq™ Hot Start version (TaKaRa, Kyoto, Japan). Sanger sequencing was performed using the Big Dye v.1.1 terminator cycle sequencing kit and an Applied Biosystems 3500xl capillary sequencer (Applied Biosystems, Foster City, CA).
2.6 Relative telomere length measurement
The relative repeat copy number of telomere (T) and single copy gene (36B4a) (S) were measured by real-time PCR in a StepOne Plus real-time PCR system (Applied Biosystems) as described previously (Cawthon, 2009). The primers for the telomere PCR were Telo-F (Forward): ACACTAAGGTTT-GGGTTTGGGTTTGGGTTTGGGTTAGTGT; Telo-R (Reverse), TGTTAGGTAT-CCCTATCCCTATCCCTATCCCTATCCCTAACA; 36B4-F (Forward), CAGCAA-GTGGGAAGGTGTAATCC; 36B4-R (Reverse), CCCATTCTATCATCAACGGGT-ACAA.
All samples for both the telomere and single copy gene amplifications were performed in triplicate in 10 μl reaction system. One reference DNA was serially diluted (twofold) with deionized water to create eight concentrations of DNA ranging from 1.0 to 128 ng/μl to determine the standard curve. The relative length of telomere was expressed as T/S ratio, reflecting the average telomere repeat copy number of each DNA sample calculated relative to the reference DNA.
2.7 Copy number variation analysis
To evaluate the copy number across the targeted genes and to identify potential large heterozygous or homozygous deletions, we analyzed the copy number of all sequenced regions of the 253 cases and 125 controls using Ion reporter 5.0.
2.8 Network analysis
Prediction of gene–gene networks for candidate genes, and their potential interactions with IPF and related phenotypes was performed using Phenolyzer, a tool for phenotype-based prioritization of candidate genes in human diseases (Yang, Robinson, & Wang, 2015). Each candidate gene was given a normalize score ranging from 0 to 1 and ranked according to their relationships with disease/phenotype and related genes (Yang et al., 2015).
2.9 Statistical analysis
Statistical analyses were carried out with the statistical program SPSS version 19.0 and results were expressed as the mean ± SD (continuous variables) and as percent totals (categorical variables). Associations for common SNPs with IPF susceptibility were evaluated by Fisher's exact test providing odds ratios (ORs), 95% confidence interval (CIs), and level of significance (P). Cumulative effect of associated alleles on the risk of IPF was estimated by ORs and 95% CIs from multivariate logistic regression analyses. The association between telomere length and age was assessed by linear regression analysis in IPF patients and age-matched controls.
We detected the associations between 647 rare nonsynonymous variants (MAF ˂ 1%) and IPF by burden test, including adaptive Sum Statistic (ASUM), cumulative minor-allele test (CMAT), and weighted sum statistic (WSS) (Han & Pan, 2010; Madsen & Browning, 2009; Zawistowski et al., 2010). These tests were performed by R software and AssotesteR package (https://github.com/gastonstat/AssotesteR). Genes with evidence for disease-associated rare variants were those with significant association (P < 0.05) by at least one burden test.
Common SNPs (MAF ≥ 0.05) were tested for Hardy–Weinberg equilibrium by Pearson's Chi-square (χ2) test. Allelic model of associations for common SNPs with IPF susceptibility were evaluated by Fisher's exact test providing ORs, 95% CIs, and level of significance (P of < 0.05).
In the cumulative analysis, the risk score of each subject was calculated as the number of risk alleles. If a subject had a single identified risk allele, the risk score was 1, and the maximum risk score was 6. Subjects who carried 0–3 risk scores were assigned to the low risk group, and those with 4–6 risk scores were assigned to the high risk group. The cumulative effect of associated alleles on the risk of IPF was estimated by ORs and 95%CIs from multivariate logistic regression analyses. A standard with P value < 0.05 was considered as significant.
3 RESULTS
3.1 Baseline characteristics
A total of 253 IPF patients and 125 matched controls were included in this study (Table 1). No significant difference was found in age (65.4 vs. 65.3 years), sexual proportion (66.8% vs. 67.2%) or tobacco use (38.7% vs. 37.6%) between the cases and controls.
| Variables | IPF cases (n = 253) | Controls (n = 125) | P value |
|---|---|---|---|
| Age (years) | 65.4 ± 11.1 | 65.3 ± 10.8 | 0.24 |
| Male (%) | 66.8 | 67.2 | 0.94 |
| Tobacco use (%) | 38.7 | 37.6 | 0.83 |
| Body mass index (kg/m2) | 23.4 ± 4.1 | 24.2 ± 3.8 | 0.31 |
| Cough (%) | 247 (97.6) | 0 | – |
| Chronic exertional dyspnea (%) | 142 (56.1) | 0 | – |
| Finger clubbing (%) | 74 (29.2) | 0 | – |
| Bibasilar inspiratory crackles (%) | 137 (54.2) | 0 | – |
| Pulmonary function test | |||
| FVC% pred | 75.2 (28.5–122.6) | – | – |
| DLCO% pred | 55.4 (15.2–85.6) | – | – |
- Age is shown in mean ± SD. For IPF cases, age means onset age. FVC % pred, percent predicted forced vital capacity; DLCO% pred, percent predicted diffusion capacity for carbon monoxide.
3.2 Targeted sequencing output
A total of 1,451 amplicon for 92 targeted genes were amplified and sequenced in 253 sporadic IPF cases and 125 healthy controls. High throughput sequencing covered 94.33% of the target region with an average base coverage depth of 776.5 folds, and 98.88% of the amplicons had at least 20 independent reads, indicating the high quality of the targeted sequencing. (Supp. Table S3)
3.3 Identification of pathogenic, LOF, likely pathogenic variants, and copy number variation
Using rigid filter criteria: (1) LOF variants (frameshift, nonsense, splicing-site, initiation codon break) with a MAF no more than 0.1% among the sequencing population, or missense variants predicted to be pathogenic or likely pathogenic by InterVar according to the ACMG guideline; (2) the ones observed in control groups were filtered out; (3) validated by Sanger sequencing, we identified two reported pathogenic variants (TERT rs121918666; TERT rs199422294), 10 LOF variants, including three frameshift insertion variants, two frameshift deletion variants, four stopgain variants, and one splicing variant (Table 2). The two RTEL1 variants were predicted to be likely pathogenic by InterVar. In total, pathogenic, LOF or likely pathogenic variants in the 92 genes were found in 4.74% (12 out of 253) of the IPF patient and six out of 12 (50%) were previously unreported. All these 12 variants were validated by Sanger sequencing and excluded from the 125 controls. Potential copy number variations were searched and further validated by real-time PCR in 92 selected genes but no large deletions were detected.
| Gene | OMIM | Type | Variant function | Variant | Novelty | Pathogenicity |
|---|---|---|---|---|---|---|
| CTC1 | 613129 | Candidate | Frameshift | NM_025099.3:c.400dupT:p.Y134fs | Novel | − |
| DTNBP1 | 607145 | Candidate | Nonsense | NM_183040.5:c.G286T:p.E96X | Novel | − |
| HPS4 | 606682 | Candidate | Frameshift | NM_152841.9:c.1087dupG:p.D363fs | Novel | − |
| LAMA3 | 600805 | Candidate | Nonsense | NM_001127717.18:c.C2116T:p.R706X | rs759225610 | − |
| MMP1 | 120353 | Candidate | Frameshift | NM_002421.7:c.988delG:p.A330fs | rs753853224 | − |
| MMP19 | 601807 | Candidate | Nonsense | NM_002429.8:c.T1155A:p.Y385X | Novel | − |
| RTEL1 | 608833 | Known | Frameshift | NM_032957.4:c.387_388del:p.T129fs | Novel | PVS1/Likely pathogenic |
| RTEL1 | 608833 | Known | Nonsense | NM_032957.34:c.C3631T:p.Q1211X | Novel | PVS1/Likely pathogenic |
| IL1RN | 147679 | Candidate | Splicing | NM_173841.3:c.74-2A > G | rs763872895 | − |
| TERT | 187270 | Known | Missense | NM_198253.10:c.G2594A:p.R865H | rs121918666 | Pathogenic |
| TERT | 187270 | Known | Missense | NM_198253.4:c.G1892A:p.R631Q | rs199422294 | Pathogenic |
| RTKN2 | – | Candidate | Frameshift | NM_145307.9:c.952dupT:p.Y318fs | rs563733406 | − |
- Novelty indicates whether the variant has been reported; PVS1 indicates the variant is regarded as an “evidence of pathogenicity very strong” variant based on the ACMG guideline; Pathogenic indicates the variant is reported as pathogenic variant in the ClinVar database.
3.4 Burden tests of rare missense variants
To reveal novel associations of selected candidate genes and IPF, we performed three kinds of burden tests (ASUM, CMAT, and WSS) of genes with identified rare missense variants in 253 IPF cases and 125 healthy controls. These tests are alternative approaches to test for associations of rare and low-frequency variant effects. In the tests, three genes were found to be statistical significant (P value < 0.05) in at least one test and had a higher burden of variants in IPF group than in control (Table 3).
| Gene name | ASUM | CMAT | WSS |
|---|---|---|---|
| CSF3R | 0.138 | 0.012 | 0.04 |
| DSP | 0.02 | 0.122 | 0.11 |
| LAMA3 | 0.004 | 0.568 | 0.356 |
- ASUM, adaptive sum test; CMAT: cumulative minor-allele test; WSS: weighted sum statistic. Shown are P values for burden tests, P < 0.05 was considered as significant, genes with no significance are not shown. CSF3R, colony stimulating factor 3 receptor; DSP, desmoplakin; LAMA3, laminin subunit alpha 3.
3.5 Risk stratification model construction using common SNPs
To identify novel SNPs associated with IPF, we performed allelic-based genetic model association analysis. Four SNPs in three genes were identified to be significantly different between IPF cases and controls (Table 4). Two SNPs, (rs3737002, rs2296160) located on complement C3b/C4b receptor 1 (CR1), are firstly revealed associations with IPF susceptibility. The other two SNPs were previously reported risk SNPs for IPF (TGFB1 rs1800470, MUC5B rs35705950).
| Gene | SNP | Allele | OR (95% CI) | P |
|---|---|---|---|---|
| CR1 | rs3737002 | C/T | 1.77 (1.28–2.45) | 0.001 |
| CR1 | rs2296160 | A/G | 1.54 (1.13–2.09) | 0.006 |
| TGFB1 | rs1800470 | G/A | 1.47 (1.08–2.00) | 0.013 |
| MUC5B | rs35705950 | G/T | 4.84 (1.12–20.94) | 0.018 |
To further construct a risk stratification mode using these identified risk SNPs, we evaluated the cumulative effects of risk scores in our study (Table 5). As the data shown, patients with more risk scores had a higher risk of IPF. Specifically, compared with individuals with 0–3 risk scores (low risk group), individuals carrying 4–6 (high risk group) had higher risk (OR = 3.47, 95%CI: 2.07–5.81, P = 2.34 × 10−6).
| Risk alleles | Control (n = 125) | IPF (n = 253) | OR (95% CI) | P value |
|---|---|---|---|---|
| 0–3 | 102 (81.6%) | 142 (56.1%) | 1.00 | – |
| 4–6 | 23 (18.4%) | 111 (43.9%) | 3.47 (2.07–5.81) | 2.34 × 10−6 |
- 0–3, low risk group; 4–6, high risk group.
3.6 Relative telomere length in IPF patients and controls
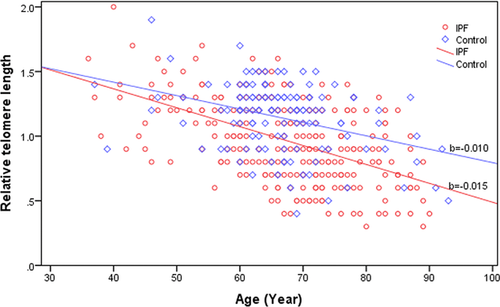
The mean telomere length of IPF patients (0.96 ± 0.32) was substantially shorter than age-matched controls (1.14 ± 0.27, P < 0.001). Telomere length distribution with age is depicted in Figure 1. The decline slope with age in telomere length in IPF patients (b = −0.015) was steeper when compared with that of the age-matched controls (b = −0.010).
3.7 Network analysis among candidate genes
Our 15 statistically significant candidate genes (from LOF located genes, burden tests identified genes, and allelic-based genetic model association analysis of SNPs identified genes) were analyzed by Phenolyzer. Network analysis of these genes showed potential gene–gene interaction and interaction with IPF-related phenotypes (Supp. Figure S2). Of these five are known IPF causal genes, and another five (ILR1, MMP1, MMP19, CSF3R, and LAMA3) were identified as significantly associated with IPF for Chinese in our study. More details are available at online mode (https://phenolyzer.wglab.org/done/56161/FEmwlynpQDNTsaIs/index.html).
4 DISCUSSION
This is the first comprehensive study using targeted resequencing (92 IPF-related genes) approach to assess the role of both common and rare variants for IPF risk in the Chinese Han population. We identified two reported FPF pathogenic variants (TERT rs121918666; TERT rs199422294), 10 LOF variants including two PVS1 likely pathogenic variants in 12/253 (4.74%) of our cohort of sporadic IPF patients, which provide potential new clues of the pathogenesis of IPF. Our study demonstrated that previously reported FPF-related genes and additional candidate genes that may also contribute to sporadic IPF cases that may, in some cases, actually be undetected familial cases. We also found that likely pathogenic variants in telomerase-related genes are still the leading genetic causes of IPF. We performed burden tests of rare variants in selected genes, and found three genes (CSF3R, DSP, LAMA3) have a statistically significant relationship with IPF. For common variants, we not only revealed four SNPs that had a statistically significant relationship with risk of IPF, but also constructed a new IPF risk-stratification model with them. In our cumulative risk model, high risk subjects had 3.47-fold (95%CI: 2.07–5.81, P = 2.34 × 10−6) risk compared with low risk subjects.
4.1 Variants in telomerase-related genes
In our study, five out of 253 (∼2%) sporadic IPF cases were identified to bear pathogenic, likely pathogenic or LOF variants in three telomerase-related genes (CTC1, RTEL1, TERT). The two likely pathogenic variants (PVS1) of RTEL1 (c.387_388del, p.T129fs; c.C3631T, p.Q1211X) are unreported novel variants.
According to previous studies, pathogenic variants of TERT and TERC occur in approximately 8%–15% of FPF and 1%–3% of sporadic IPF patients (Armanios et al., 2007; Tsakiri et al., 2007). Shorter telomere lengths are found in and are associated with decreased survival for IPF patients (Dai et al., 2015; Stuart et al., 2014). In addition, recent studies showed that rare variants in regulator of telomere elongation helicase 1 (RTEL1) are also involved in the telomere shortening and FPF (Alder et al., 2015; Hisata et al., 2013; Kannengiesser, Borie, & Revy, 2014; Kropski et al., 2014; Stuart et al., 2015). Another telomerase-related gene is CST telomere replication complex component 1 (CTC1), a causal gene of dyskeratosis congenita—a rare genetic disorder may occur pulmonary fibrosis and shorter telomere lengths (Mason & Bessler, 2011). We report here a CTC1 frameshift insertion (c.400dupT, p. Y134fs) in a Chinese sporadic IPF patient.
4.2 Variants in collagen and extracellular matrix related genes
Current knowledge of IPF pathogenesis suggests that genetic factors trigger repetitive epithelial cell injury, abnormal repair responses and matrix accumulation, and subsequently leads to progressive fibrosis and loss of lung function (Puglisi, Torrisi, Giuliano, Vindigni, & Vancheri, 2016). Excess matrix accumulation is thought to be an important part in the pathological process of IPF, and a related protein such as MMP1 that is strongly upregulated in IPF is proposed to be a potential peripheral blood biomarker (Rosas et al., 2008). In addition, a case–control study found that polymorphisms in the MMP1 promoter may confer increased risk for IPF (Checa et al., 2008). A knockout mice model of another related gene MMP19 showed a significantly increased lung fibrotic response to bleomycin compared with WT mice (Yu et al., 2012). Based on these observations, we hypothesize that variants of collagen and extracellular matrix related genes may also have potential relevance to IPF.
Our study identified a frameshift deletion (c.988delG, p.A330fs) in MMP1 and a stop gain (c.T1155A, p.Y385X) in MMP19, respectively. Interestingly, we found that our youngest patient, with an onset at 35 years of age, carried three potential functional variants in related genes (MMP1, c.988delG, p.A330fs, LOF; ITGA3, c.C469T, p.R157C, rs557579280; MMP19, c.C712T, p.R238W, rs754912368). These two missense variants were extremely rare in the ExAC database and they were predicted in silico to change the protein function. These findings suggest that interactions between multiple variants may predispose to IPF. Functional studies are needed for confirmation.
4.3 Variants in HPS-related genes
Hermansky-Pudlak syndrome is a heterogeneous and rare autosomal recessive genetic disorder (El-Chemaly & Young, 2016; Vicary et al., 2016). Patients with HPS-1, HPS-2, and HPS-4 tend to develop pulmonary fibrosis (Vicary et al., 2016). We included HPS-related genes in our study, and identified one novel frameshift insertion in HPS4, and one stop gain variant in DTNBP1 (HPS7) (Table 2). The IPF patients who carried these candidate variants were carefully examined and findings of HPS were not seen. We propose that some subtypes of IPF and HPS may result from the same genetic factors.
4.4 Correlations between phenotype and genotype
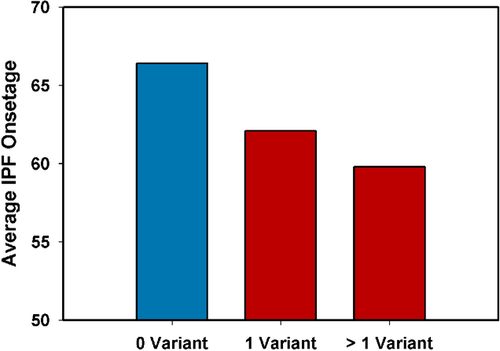
Thus far at least 13 genes are known to cause IPF but their genotype–phenotype correlation is poorly understood. We firstly found candidate variants known to cause specific syndromes with lung fibrosis (CTC1, Dyskeratosis Congenita; DTNBP1, HPS, and HPS4, HPS) that may also contribute to non-syndromic sporadic IPF pathogenesis. We did not find signs or symptoms of these disorders in our IPF patients, despite careful examination. We compared the average onset age of our IPF cases with the number of identified variants (pathogenic, likely pathogenic and LOF) (Figure 2). Our results suggest that IPF cases who carried more variants had an earlier average onset age.
4.5 Common SNPs and cumulated risk analysis for IPF
Previous studies indicate that the gain-of-function promoter variant (MUC5B, rs35705950) is associated with both FPF and sporadic IPF in different populations (Horimasu et al., 2015; Lee & Lee, 2015; Peljto et al., 2013; Seibold et al., 2011). In our study, the frequencies of the high risk T allele were 3.70% and 0.80% in IPF patients and healthy controls respectively (P = 0.018), similar to previous results in Chinese and no TT homozygote was detected (Wang et al., 2014). Considering that IPF is a highly heterogeneous and complex disease, we performed cumulated risk analysis of four significant common SNPs (rs3737002, rs2296160, rs1800470, and rs35705950). In the analysis, subjects who carried 4–6 risk scores (high risk group) had a 3.47-fold increased risk compared with subjects who carried 0–3 risk scores.
Although our study identified several pathogenic, likely pathogenic and LOF variants in 253 sporadic IPF patients in Chinese population, it has several limitations. First, only 92 candidate IPF genes were included and whole exome sequencing was not done. Second, we only studied pathogenic, likely pathogenic and LOF variants that were in exon or short exonic flanking regions, and synonymous variants and variants in intronic or untranslated regions were not studied. Thus, synonymous and noncoding variants are likely underestimated. Third, functional studies of our candidate variants are needed to confirm their causative role.
In conclusion, we report here the first study of the role of both rare and common variants for IPF risk in the Chinese population. Our study identified multiple novel rare and common variants that are associated with increased risk of IPF. Our cumulative risk model analysis results suggest the possibility of risk prediction and stratification for IPF in Chinese.
ACKNOWLEDGMENTS
We would like to acknowledge all the participants who volunteered for this study.
DISCLOSURE STATEMENT
The authors declare no conflict of interest. All variants in Table 2 have been submitted to the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/).