A PIGH mutation leading to GPI deficiency is associated with developmental delay and autism
Communicated by Garry R. Cutting
Abstract
We identified an individual with a homozygous missense variant (p.Ser103Pro) in a conserved residue of the glycosylphosphatidylinositol (GPI) biosynthesis gene PIGH. This gene encodes an essential component of the phosphatidylinositol N-acetylglucosaminyltransferase complex, in the first step of the biosynthesis of GPI, a glycolipid anchor added to more than one hundred human proteins, several being critical for embryogenesis and neurological functions. The affected individual had hypotonia, moderate developmental delay, and autism. Unlike other reported individuals with GPI deficiency, the proband did not have epilepsy; however, he did have two episodes of febrile seizures. He had normal alkaline phosphatase and no brachytelephalangy. Upon analysis of the surface expression of GPI-anchored proteins on granulocytes, he was demonstrated to have GPI deficiency. This suggests that PIGH mutations may cause a syndrome with developmental delay and autism, but without an epileptic encephalopathy, and should increase the awareness of the potentially deleterious nature of biallelic variants in this gene.
Glycosylphosphatidylinositol (GPI) is a glycolipid that anchors over 150 proteins to the cell surface, these proteins are important for embryogenesis, neurogenesis, and signal transduction. More than 30 genes involved in the biosynthesis of these GPI anchors have been identified to date (Kinoshita, 2016), and over 20 of them have been associated with human diseases. Mutations in GPI biosynthesis genes result in a wide spectrum of symptoms, but usually include global developmental delay, intellectual disability, and epilepsy. In this report, we study an individual with a homozygous variant in PIGH (MIM# 600154), a GPI biosynthesis gene that has never been previously associated with human diseases.
The proband is the only child of a consanguineous couple of Indian origin (second cousins). There had been a previous miscarriage following placental abruption at 6 months gestation. The proband was born at term following an emergency C-section due to nonreassuring fetal heart tracing. His birth weight was 2.95 kg, and his head circumference was 33 cm. He had two episodes of febrile seizures at the age of 17 months, but these did not recur; he did not require antiepileptic drugs, and EEG and brain MRI were normal. He sat without support at 8 months, walked at 13 months and began using occasional words at 2 years old. He was diagnosed with hypotonia, moderate global developmental delay and autism at 2.5 years old.
He was last assessed at 5 years 2 months, his weight was 19.5 kg (59th percentile) and height was 113.0 cm (74th percentile), whereas his head circumference was 49.5 cm (12th percentile, −1.15 SD). He was not dysmorphic but has a high palate and slight hirsutism. He did not have abnormal digits or nails on the hands and feet. He has echolalic speech and limited spontaneous words and phrases that he uses to label objects. He is able to count from 1 to 10, but does not know the value of numbers; he can say the alphabet and identify some colors and shapes. He sleeps well and eats well. He is able to finger feed but does not yet use utensils. He is not yet toilet trained and needs assistance with dressing and undressing. He has difficulty with coordinated skills such as jumping and catching a ball. Investigations revealed a low ferritin, but normal total iron binding capacity. There were normal plasma amino acids, carnitine, as well as urine organic acids, guanidinoacetate, and creatinine. A plasma acylcarnitine profile showed mildly increased levels of C8, C10:1, and C10, but with normal C8/C10 ratio. Serum alkaline phosphatase was normal at 227 U/l (range is 100–320 U/l).
Trio whole exome sequencing was performed at GeneDx. Briefly, using genomic DNA from the proband and parents, the exonic regions and flanking splice junctions of the genome were captured using the Clinical Research Exome kit (Agilent Technologies, Santa Clara, CA). Massively parallel (NextGen) sequencing was done on an Illumina system with 100 bp or greater paired-end reads. Reads were aligned to human genome build GRCh37/UCSC hg19, and analyzed for sequence variants using a custom-developed analysis tool. Additional sequencing technology and variant interpretation protocol has been previously described (Tanaka et al., 2015). The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page (https://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/).
A homozygous missense variant was identified in PIGH (NM_004569.3:c.307T > C, p.Ser103Pro). No other candidate variant was identified on the exome sequencing. Given that variants in other components of the GPI biosynthesis pathway had previously been identified in individuals with developmental delay and other features, this variant was considered to be a good candidate gene to explain the delays in this individual. Moreover, the variant is very rare (rs776038451, MAF of 0.00001625 with four heterozygotes identified in gnomAD, no homozygotes), affects a residue highly conserved in other organisms (see Figure 1), and changes it for a proline which would significantly alter the protein structure (Bajaj et al., 2007). The variant has a CADD score of 27 which means that it is in the highest 1% of most deleterious variants in the genome, and is also predicted to be deleterious by SIFT (score 0.024, cutoff ≤0.05), and possibly damaging by Polyphen2 (HDIV score 0.896 (range for possibly damaging 0.453–0.956) and HVAR score 0.635 (range for possibly damaging 0.447–0.909)). The scores were obtained using wANNOVAR (Yang & Wang, 2015), which uses dbNSFP v3.0 (Liu, Wu, Li, & Boerwinkle, 2016). The variant has been entered in the LOVD mutation database for PIGH (https://www.lovd.nl/PIGH). The patient was enrolled after informed consent from the family in a research protocol approved by the Institutional Review Board of the CHU Sainte-Justine Research Center. We then examined whether the cell surface levels of GPI-AP were altered. As shown in Figure 1, CD16 at the cell surface of granulocytes was decreased by up to 51% when compared with both parents, who are asymptomatic, and each carry a heterozygous mutation. This result thus demonstrates that homozygosity of the Ser103Pro leads to a decreased ability to synthesize GPI anchored proteins.
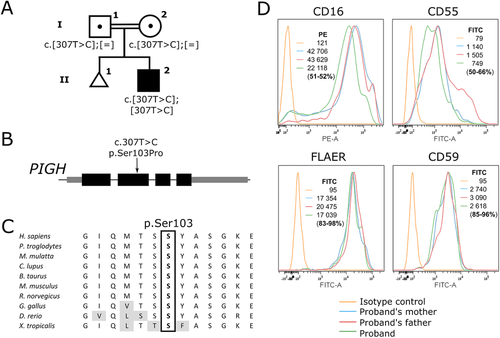
The gene encoding phosphatidylinositol glycan anchor biosynthesis class H (PIGH) was first cloned by Kamitani, Chang, Rollins, Waneck, and Yeh (1993). PIGH is an essential component of the GPI–N-Acetylglucosaminyltransferase (GPI–GlcNAc transferase) complex, also composed of the proteins PIGA, C, P, Q, and Y (Doering, Masterson, Englund, & Hart, 1989; Kinoshita, Inoue, & Murakami, 2014). Human diseases have been associated with mutations in other proteins of the complex. Acquired somatic mutations in the X-linked PIGA gene cause paroxysmal nocturnal hemoglobinuria (Takeda et al., 1993), characterized by complement-mediated destruction of red blood cells due to a loss of the protective GPI-anchored proteins CD55 and CD59 (Brodsky, 2008). Inherited mutations in PIGA cause an especially severe GPI-deficiency disorder, with half of the reported individuals passing away at a young age of liver failure and pneumonias following a severe epileptic encephalopathy. The phenotype can be milder for mutations in other genes encoding members of this complex. Of note, two siblings with mutations in PIGY (out of the four reported individuals to date) did not present with seizures or abnormal digit, and had normal alkaline phosphatase (Ilkovski et al., 2015; Knaus et al., 2018). Therefore, these individuals and the proband described here demonstrate how deleterious mutations in the GPI–GlcNAc transferase complex can lead to relatively milder phenotypes compared with other inherited GPI deficiency disorders. Thus, an increased index of suspicion is warranted for GPI-deficiency disorders, especially since in some cases, the epilepsy can respond to B6 supplementation (Kuki et al., 2013; Thompson et al., 2006).
ACKNOWLEDGMENTS
We acknowledge funding by the Canadian Institutes for Health Research (clinician-scientist award RN315908 to P.M.C.), the Fonds de Recherche du Québec – Santé (FRQS) clinical research scholar award to P.M.C., and the Fondation du Grand Défi Pierre Lavoie.
DISCLOSURE STATEMENT
The authors declare no conflict of interest.