Mediterranean Founder Mutation Database (MFMD): Taking Advantage from Founder Mutations in Genetics Diagnosis, Genetic Diversity and Migration History of the Mediterranean Population
Communicated by Alastair F. Brown
ABSTRACT
The Mediterranean basin has been the theater of migration crossroads followed by settlement of several societies and cultures in prehistoric and historical times, with important consequences on genetic and genomic determinisms. Here, we present the Mediterranean Founder Mutation Database (MFMD), established to offer web-based access to founder mutation information in the Mediterranean population. Mutation data were collected from the literature and other online resources and systematically reviewed and assembled into this database. The information provided for each founder mutation includes DNA change, amino-acid change, mutation type and mutation effect, as well as mutation frequency and coalescence time when available. Currently, the database contains 383 founder mutations found in 210 genes related to 219 diseases. We believe that MFMD will help scientists and physicians to design more rapid and less expensive genetic diagnostic tests. Moreover, the coalescence time of founder mutations gives an overview about the migration history of the Mediterranean population. MFMD can be publicly accessed from http://mfmd.pasteur.ma.
INTRODUCTION
The Mediterranean Basin is the region of lands that surround the Mediterranean Sea at the intersection between three different continents: Africa, Asia and Europe. Since the original diaspora of peoples from the African continent (Capelli et al. 2006), its population history witnessed many civilization movements both in prehistorical and historical times, as it was and still is an important route of transport, trade and interaction. The Neolithic period is marked by the transition from hunter-gathering to the spread of agriculture, that led to a demographic transition, by increasing the population growth rate (Bocquet-Appel and Bar-Yosef 2008). In addition, the Neolithic period is marked by the development of trading between different civilizations and by several migratory movements. Ancient civilizations were located around the Basin, including, the Phoenician, the Greek, the Roman and the Arab ones. The Phoenicians used their maritime expertise to become the principal traders in the Mediterranean Sea, as they spread throughout the Mediterranean basin and establish important colonies and centers of trade (Zalloua et al. 2008). Indeed, starting from Carthage, the most important Phoenician colony, this population spread to settle throughout the western Mediterranean regions of the North African coast and the Iberian Peninsula (Abulafia 2011). The Greeks had multiple colonies along the western Mediterranean and Black Sea coasts, many of these being in Italy. They contested with the Phoenicians over control of colonies and trading posts in Sicily and the western Mediterranean (Sacks and Murray 2009). After the Third Punic War between the Romans and Phoenicians, the Roman Empire became the most powerful force in the Mediterranean region (Abulafia 2011). Then, in the early part of the middle ages, the Arabs conquered a large region of the Mediterranean Sea, unifying the Arabian Peninsula with a vast part of the Byzantine Empire, the whole Persian lands, North Africa and the Iberian Peninsula (Hitti 1948).
All these demographic events, population interactions and migratory movements that occurred along the Mediterranean Sea, had a crucial impact on the genetic structure of the Mediterranean population, which have already been studied using Mitochondrial DNA (mtDNA), Y chromosome and Autosomal molecular markers and sequencing. The population history and diversity have important medical genetic implications (Laberge et al. 2005). In particular populations, many disease-causing mutations can be traced to a founder individual, in whom the mutation first appeared. Founder effect occurs when a new group is established by a small number of individuals that are separated from a larger population. The new population represents only a restricted sample of the genetic variability that was present in the original population (Neuhausen 2000) and further endogamy led to modifications in DNA variation frequencies. Therefore, founder effects explain the high prevalence of some disease-causing mutations in a particular population, and haplotype analysis can demonstrate that a common mutation is due to a founder effect with linkage disequilibrium to the adjacent genetic markers. The size of the linkage disequilibrium intervals helps to estimate the age of the original mutation spreading into a given population (Zeegers et al. 2004). Identification of founder mutations in a particular population has been useful for the improvement of molecular diagnosis and genetic counseling. Screening for few prevalent founder mutations is quicker and cheaper than testing many rare mutations (Ferla et al. 2007). Moreover, determining founder mutations and their regional distribution in different ethnic groups can help scientists to study the genetic diversity and migration history of these populations (Ostrer 2001).
There are a lot of published findings that highlight the importance of founder mutations in medical genetics. For instance, various ancient founder mutations transmitted across tens or hundreds of generations, tend to migrate over long distances and spread widely around the Mediterranean Sea. This case is illustrated by the c.35delG common founder mutation in the GJB2 gene that causes deafness, and appears to have arisen in the Middle East about 500 generations ago (10000 years) and spread across Europe and around the Mediterranean basin (Van Laer et al. 2001). Furthermore, several genetic disorders have a wide geographical presence along the Mediterranean region, with multiple disease-causing founder mutations. This situation is observed for the familial Mediterranean fever disease; multiple founder mutations affecting the MEFV gene were shared between different Mediterranean populations. Phylogenetic analyses based on allele frequencies of the most common MEFV founder mutations (M694V, V726A, M680I, M694I, and E148Q) in 14 Mediterranean populations, suggested three major possible gene flows between populations (Papadopoulos et al. 2008). The current distribution pattern of MEFV mutations may be influenced by several historical events: the Byzantine Empire, the Arab conquests, the Ottoman dominance, the dispersal of the Armenian nation and the Jewish Diaspora (Papadopoulos et al. 2008). Screening frequent founder mutations in a particular population as the first step in molecular genetic diagnosis is a cost-effective methodology before considering alternative approaches such as Next Generation Sequencing (Ponti et al. 2015).
In this paper, we describe the Mediterranean Founder Mutation Database (MFMD), a comprehensive online database established to collect and document human founder mutations reported in the Mediterranean population. We believe that this is the first database for founder mutations in the Mediterranean region. The database will help scientists and physicians in having an overview about the spectrum of founder mutations found in the Mediterranean population, and will be beneficial to understand the history, demography and migration flows of the Mediterranean populations. Furthermore, MFMD provides useful information for the diagnosis and prevention of genetic diseases. The MFMD can be publicly accessed from http://mfmd.pasteur.ma.
METHODS
Software design and implementation
The MFMD was implemented using a three-tier model (client, application server and database). The web interface was designed following the Model-View-Controller (MVC) design pattern, and developed using HTML (HyperText Markup Language), CSS (Cascading Style Sheets) and Javascript programming languages. The PHP (version 5.4) scripting language was used for all data retrieval and output. The data were managed with MySQL (version 5.5) database management system. The PHPlot 6.1.0 tool (http://phplot.sourceforge.net/) was used to generate graphs in the statistics page of the MFMD web interface. The R statistical programming language was used to draw Bar charts and to perform Heatmap Analysis.
Literature search and data extraction
We conducted a literature search using the PubMed database for articles, published before March, 1st 2015. The search strategy to identify all relevant studies was based on a combination of the following keywords: (“founder” OR “common”) AND (“effect” OR “mutation” OR “allele” OR “variation” OR “variant” OR “loci” OR “locus” OR “haplotype” OR “microsatellite” OR “polymorphism”), without any restriction on language. In addition, to determine articles conducted in subjects of Mediterranean origin, we used a list of Mediterranean country names to filter out non relevant studies. Remaining abstracts were downloaded in XML-format from PubMed and stored in the database. Data were extracted from abstracts by independent reviewers and validated by the database editor in chief. Genes, diseases and mutations were identified and highlighted in abstracts using the PubTator text mining tool, to facilitate data extraction. Eligible studies were selected if they were conducted in subjects of Mediterranean origin, and reported a founder mutation. We collected both founder mutations proved by haplotype analysis and mutations with suggested founder effect. Moreover, we also checked all references cited in the identified articles for any additional literature. The following data were extracted from the eligible studies: publication information, subject information (population of origin, geographical region and ethnic group) and mutation information (DNA change, amino acid change, prevalence of mutation among affected individuals and coalescence time) (Figure 1A). The Mutalyzer 2.0.6 software (Wildeman et al. 2008) was used to check mutation nomenclature according to the HGVS recommendations. Gene symbol follows the HUGO gene nomenclature (Gray et al. 2014). Genetic diseases registered in the database have been classified using the World Health Organization (WHO) International Statistical Classification of Diseases and Related Health Problems 10th Revision (ICD-10) Version 2010 (http://apps.who.int/classifications/icd10/browse/2010/en).
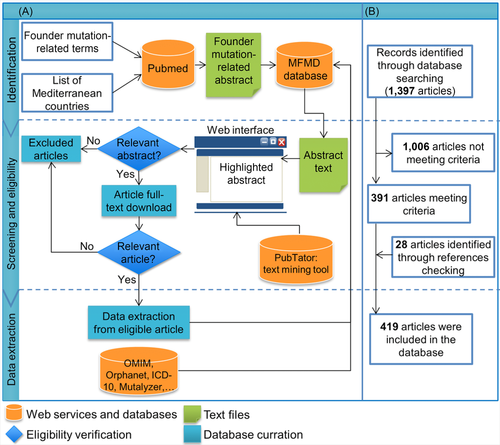
The current version of the database registered founder mutation data from the following Mediterranean countries: Albania, Algeria, Bosnia and Herzegovina, Croatia, Cyprus, Egypt, France, Greece, Israel, Italy, Lebanon, Libya, Malta, Monaco, Morocco, Palestine, Slovenia, Spain, Syria, Tunisia and Turkey. All patients are classified according to their population of origin before emigration.
RESULTS
Data included in the database
The flowchart of the study selection is shown in Figure 1B. The literature search yielded 1397 publications. A total of 1006 studies did not meet the inclusion criteria and were rejected based on the title, abstract or the full text screening. Three hundred and eighty-one studies met the inclusion criteria and were included in the database. Moreover, 28 additional articles were obtained from reviewing references from the eligible studies. Finally, a total of 419 studies were included in the database. These relevant articles dated from 1989 to 2015 and the distribution of number of articles per year showed an increasing trend (Figure 2).

The MFMD web interface organization and functionality
The web interface allows several query options to search mutation data cataloged in the MFMD database. To retrieve the corresponding information of the founder mutation of interest, the website provides two ways to query the database (Figure 3). Users can use the search box to explore the content of the database by keywords related to disease name, gene symbol, OMIM ID and patient origin. In addition, the single letter search option arranges diseases and genes in alphabetical order. Clicking on any letter will provide all available diseases and genes starting with this letter. When the user selects a disease or gene of interest from the search result, the related mutation data will be displayed. For each mutation, a summary of information is provided including DNA change, protein change, location, reference sequence ID, mutation type and effect. In addition, mutation frequency among affected individuals and mutation coalescence time are provided when available. The gene record includes: gene symbol, gene full name and chromosome location. For each disease entry, details such as phenotype name, disease classification according to the World Health Organization International Classification of Disease (WHO-ICD) and the mode of inheritance are provided. Moreover, links to online resources are available to get further detailed information about a specific disease or gene, including OMIM, Orphanet, Genetic testing registry, ClinicalTrials, Genome Browser, Ensembl, UniProt GeneCards, Kyoto Encyclopedia, BioGPS, HGNC and HPRD.
Characteristics of disease and gene data
A total of 219 Mendelian diseases are registered in MFMD. Data analysis reveals that 67.1% of all the inherited diseases follow autosomal recessive mode of inheritance, while autosomal dominant transmission represents 21.9% of all cases. In addition, 2.3% are X-linked and 8.7% of genetic diseases have multiple modes of transmission (Figure 4A). According to the WHO-ICD-10 disease classification, the main disease categories in the database include endocrine, nutritional and metabolic diseases (26.5%), followed by nervous system diseases (20.5%) and congenital malformations and chromosomal abnormalities (17.4%). Diseases of blood and immune mechanism (11%), neoplasms (7.3%), diseases of the eye and adnexa (5.5%) are also prevalent (Figure 4B).

Comparison of founder mutation spectrum between Mediterranean populations
At the time of its first online release, the MFMD database contained 383 founder mutations. At present, the disease with the highest number of founder mutations is Breast-ovarian cancer, with 21 mutations in the BRCA1 gene and 13 mutations in the BRCA2 gene, the majority of them being found in Spanish, Italian and Greek populations. Founder mutations are also prevalent in Ataxia-telangiectasia disease, with 15 out of 18 mutations in the ATM gene reported in the Italian population. The 35delG mutation in GJB2 gene is the most common mutation, detected in 8 Mediterranean countries from Europe (Greece, Italy and Spain), Asia (Lebanon and Turkey), and Africa (Egypt, Morocco and Tunisia). More than one founder mutation is responsible for the Beta thalassemia in the Mediterranean population. The most recurrent mutation c.118C>T (also known as Codon 39, C>T) in HBB gene has been reported in 3 populations. Five founder mutations responsible for Familial Mediterranean fever widely spread around the Mediterranean Sea, were described in 4 populations (Egypt, Lebanon, Syria and Tunisia). On the other hand, current data showed that 270 mutations were reported in only one population.
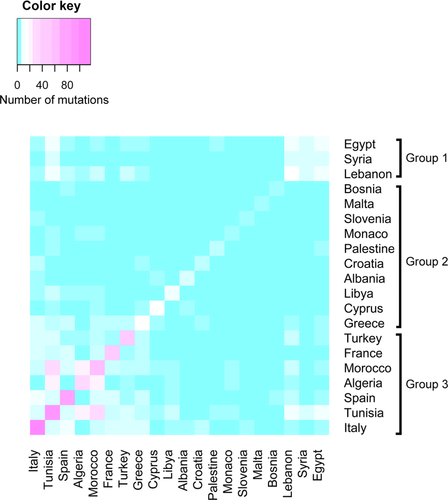
We clustered Mediterranean populations, using the number of shared mutations as a linkage criterion which indicates the similarity between populations. The results are presented as a heatmap in Figure 5. Each colored cell of the Heat Map indicates the number of founder mutations shared between two countries, and the diagonal cells show the total number of mutations in a particular country. Current data indicate the presence of more than 117 founder mutations in the Italian population, which represents 30.55 % of all mutations included in the database. Italy is followed by Tunisia, Spain and Morocco with 89, 71 and 54 mutations, respectively. Based on the resulting pair wise comparisons between populations, we can define three clusters: the first is composed of 3 Arab populations, including Egypt, Lebanon and Syria. The second is mainly composed of European populations (Albania, Bosnia-Herzegovina, Croatia, Cyprus and Greece, Malta, Monaco and Slovenia) and contains a small number of founder mutations. The third group was composed of 4 European populations (France, Italy, Spain and Turkey) and 3 North African populations (Algeria, Morocco and Tunisia).
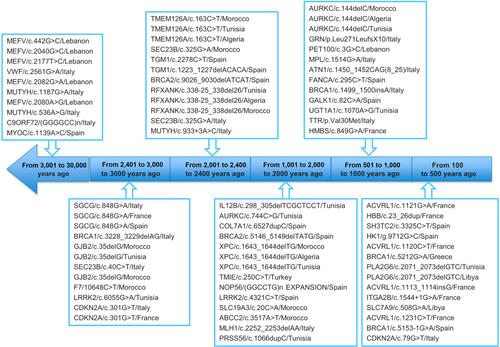
Coalescence times of several founder mutations have been determined in Mediterranean populations; they are shown in Figure 6 together with the gene symbol and population in which the mutation occurred. There are 60 mutations in the database with known age, their coalescence time ranging from 30000 to 100 years ago. The three oldest mutations were found in the MEFV gene with the following coalescence time: 30000, 23000 and 15000 years ago. The most recent founder effect was reported in the ACVRL1 gene and occurred 100 years ago.
DISCUSSION
The Mediterranean basin has been the point of intersection between several civilizations, and religions that influenced each other (Abulafia 2011), thus providing an exceptional context to examine the impact of demographic events and migration on the actual genetic landscape. The study of the genetic structure of Mediterranean populations using Y-chromosome microsatellites (Quintana-Murci et al. 2003) and mtDNA variations (Plaza et al. 2003), revealed a low genetic structure between the northern and southern shores of the Mediterranean sea. In addition, Y-chromosome analysis failed to find any significant barrier to genetic exchange between populations from the eastern part of the Mediterranean (Manni et al. 2002). On the basis of X-chromosome polymorphisms, high overall genetic population homogeneity was found in the Mediterranean area, except in north-west African populations (Morocco) (Tomas et al. 2008). In the western part of the Mediterranean area, the Strait of Gibraltar has been suggested by several studies as a geographic barrier to gene flow between north-west Africa and the Iberian Peninsula (Tomas et al. 2008)(Ennafaa et al. 2009). The Mediterranean Sea might also act as a partial barrier to gene flow between the northern and southern shores (Athanasiadis et al. 2010). On the other hand, the Arab region played an important role in shaping the current genetic structure of the Mediterranean population. It was an important area for human population interactions and movements, as well as an interesting bridge for gene flow between Africa, Asia, and Europe (Tadmouri et al. 2014).
MFMD is a carefully curated database extracted from the literature for all described founder mutations reported in the Mediterranean population, providing detailed information for each one. A user-friendly web interface was designed to provide a flexible access to the database content. MFMD could be a useful resource for research purposes in medical and population genetics studies on the Mediterranean population.
More than 30% of founder mutations registered in the database were reported in the Italian population; several of them have been found in Sardinia and Sicily islands. Due to geographical and cultural barriers to gene flow, some populations can be considered as genetic isolates which lack significant genetic sub-structures such as Sardinian (Pardo et al. 2012; Di Gaetano et al. 2014) and Sicilian populations (Rickards et al. 1998; Sarno et al. 2014). The Italian Peninsula hosted various human migration events and cultural exchanges, due to its central geographical location in the Mediterranean basin. Furthermore, numerous ethno-linguistic minorities inhabit Italy and represent 5% of the total population (Capocasa et al. 2014). Consequently, the combination of linguistic and geographic factors may have influenced the genetic isolation of Italian populations (Capocasa et al. 2014) in combination with endogamy and genetic drift phenomena.
The database registered 60 mutations with known coalescence time, 24 older than 2000 years ago, and 36 mutations more recent. We observed that 13 out of 24 old mutations were reported in more than two populations; in contrast only 6 out of 36 recent mutations were reported in different populations. Thus, we can hypothesize that founder mutations reported in only one population, which is the case of 270 mutations, can be due to recent founder effects.
The Heat Map constructed based on the number of shared mutations between Mediterranean populations, revealed three main groups (labelled as Group 1, Group 2 and Group 3). The number of founder mutations reported in populations belonging to Group 1 and Group 2 was relatively low. Both groups were mainly composed of Eastern Mediterranean countries, for which insufficient amount of molecular data on genetic diseases has been published. The interest for human genetics research in this part of the Mediterranean region is relatively recent and there remains a clear need for more investments to build capacities in medical genetics. In contrast, remarkable efforts in exploring the molecular bases of genetic disorders in western Mediterranean countries forming Group 3, has led to a wide characterization of disease-causing mutations. Clinicians and researchers from North Africa (Morocco, Tunisia and Algeria) benefit from collaboration with northern Mediterranean partners to bridge the gap in healthcare infrastructures and to overcome the lack of molecular genetic technologies. The Heat Map showed that western Mediterranean populations (Morocco, Algeria, Tunisia, France, Italy and Spain) hold the majority of founder mutations and have several shared founder mutations. In particular, North African populations (Morocco, Algeria and Tunisia) shared 15 founder mutations. This is in agreement with the analysis of mitochondrial DNA (mtDNA) sequences performed in Western Mediterranean populations that showed multidirectional gene flow in this region (Plaza et al. 2003). Moreover, recent migration events from North Africa have a significant contribution to the genetic diversity in the southern European region, consequently North African disease risk alleles should be taken in consideration when developing molecular diagnostic tests for genetic disorders in European populations (Botigué et al. 2013).
Differences in the prevalence of many diseases and mutations have been observed in different populations and ethnic groups, as a result of founder effects (Neuhausen 2000). For example, more than 30 inherited diseases are more prevalent in the Finland than in other populations (Polvi et al. 2013). On the other hand, many mutations in MFMD database are unique to a specific ethnicity, such as Jewish groups. Thus, patient ancestry can be used as a criterion for diagnosis (Ostrer 2001).
To date, the database contains 219 genetic diseases; the majority of them follow an autosomal recessive mode of inheritance (67.1%). The high rate of consanguinity in this region and especially in Arab countries may contribute to this observation (Tadmouri et al. 2009). The disease classification reveals that the majority of diseases in MFMD belong to three categories: endocrine, nutritional and metabolic diseases (26%), neurodegenerative diseases (21%) and congenital malformations and chromosomal abnormalities (17%). Similarly, endocrine and metabolic diseases are also prevalent in Morocco; they represent 24% of all genetic disorders reported in the Moroccan Genetics Disease Database (MGDD), then followed by congenital malformations and chromosomal abnormalities (22%) and nervous system diseases (19%) (Charoute et al. 2014). In contrast, the classification of genetics disease registered in the “Catalogue of Transmission Genetics in Arabs" (CTGA) database, showed that congenital malformations and chromosomal abnormalities are the most prevalent category of genetic disorders in the Arab population (the United Arab Emirates, Bahrain, Oman and Qatar). They represent 34% of all recorded diseases, followed by endocrine and metabolic disorders (19%) and nervous system disorders (11%) (Tadmouri 2012). Moreover, the classification of genetic diseases reported in Tunisia is in accordance with the data observed in the Arab population. Congenital malformations and chromosomal abnormalities constitute 30% of all genetic disorders found in the Tunisian population, followed by endocrine and metabolic disorders (19%) and nervous system disorders (11%) (Romdhane et al. 2011). Differences in the classification of genetic disorders may be explained by the fact that genetic disorders are not equally distributed in the Arab region, where some genetic diseases are presents in many countries, whereas almost half of diseases were reported only in a single population (Tadmouri et al. 2014). In addition, the majority of MFMD data were collected from European countries (Italy, France and Spain), consequently the heterogeneity between populations registered in both databases (MFMD and CTGA) may be responsible for these differences in the classification diseases.
Founder mutation data were collected from 419 articles, the distribution of these studies against time shows an increasing trend. The number of founder mutations discovered in the Mediterranean population is expected to keep increasing in the coming years, due to the use of next-generation sequencing technology. In this respect, identification of founder mutations facilitates the development of efficient molecular diagnostic assays. Testing prevalent founder mutations would be cheaper and quicker than screening for various rare mutations. In addition, the number of carriers of a given founder mutation, allows scientists to perform penetrance analyses of this mutation among populations (Neuhausen 2000; Zeegers et al. 2004; Ferla et al. 2007; Romdhane et al. 2012).
In conclusion, we developed an online database of founder mutations in the Mediterranean population. It provides relevant information to the research community, through a user-friendly interface. All efforts have been made to construct a comprehensive database by conducting an exhaustive search for relevant studies reporting founder mutations. However, the current version of the MFMD database might miss some mutation data. To ensure the collection of the complete set of founder mutation data in the Mediterranean population, we will develop new research strategies including more biomedical literature databases and text-mining tools. We will continue collecting new founder mutation data from published literature, to keep MFMD updated. In addition, the database supports new data submission from public users, using an online submission form. The MFMD database will be beneficial to the improvement of disease diagnosis and better understanding the gene flow in the Mediterranean population.
ACKNOWLEDGMENTS
This work was supported by Pasteur Institute of Morocco (IPM) and a collaborative project between the French National Institute of Health and Medical Research (INSERM) and the Moroccan National Centre for Scientific and Technical Research (CNRST).




