Activating Mutations Affecting the Dbl Homology Domain of SOS2 Cause Noonan Syndrome
Communicated by Nancy Spinner
Correspondence to: Benjamin G. Neel, The Laura and Isaac Perlmutter Cancer Center, New York University School of Medicine, New York, NY 10016. E-mail: [email protected]
Correspondence to: Marco Tartaglia, Department of Hematology, Oncology and Molecular Medicine, Istituto Superiore di Sanità, Viale Regina Elena, 299, Rome 00161, Italy. E-mail: [email protected]
Contract grant sponsors: National Institutes of Health (R01 HL071207; R01 HL0832732; U01 DE020060, R01 HG003988, and U54 HG006997; U54 HG006504); Telethon-Italy (GGP13107); AIRC (IG 13360); Ministry of Health (RF-2011-02349938); Princess Margaret Cancer Foundation and the Ontario Ministry of Health and Long Term Care; CIHR CGS-D.
ABSTRACT
The RASopathies constitute a family of autosomal-dominant disorders whose major features include facial dysmorphism, cardiac defects, reduced postnatal growth, variable cognitive deficits, ectodermal and skeletal anomalies, and susceptibility to certain malignancies. Noonan syndrome (NS), the commonest RASopathy, is genetically heterogeneous and caused by functional dysregulation of signal transducers and regulatory proteins with roles in the RAS/extracellular signal-regulated kinase (ERK) signal transduction pathway. Mutations in known disease genes account for approximately 80% of affected individuals. Here, we report that missense mutations altering Son of Sevenless, Drosophila, homolog 2 (SOS2), which encodes a RAS guanine nucleotide exchange factor, occur in a small percentage of subjects with NS. Four missense mutations were identified in five unrelated sporadic cases and families transmitting NS. Disease-causing mutations affected three conserved residues located in the Dbl homology (DH) domain, of which two are directly involved in the intramolecular binding network maintaining SOS2 in its autoinhibited conformation. All mutations were found to promote enhanced signaling from RAS to ERK. Similar to NS-causing SOS1 mutations, the phenotype associated with SOS2 defects is characterized by normal development and growth, as well as marked ectodermal involvement. Unlike SOS1 mutations, however, those in SOS2 are restricted to the DH domain.
Introduction
Aberrant signaling from RAS has been causally linked to a family of clinically related developmental disorders, collectively termed the RASopathies, which are characterized by facial dysmorphism, a wide spectrum of cardiac defects, reduced growth postnatally, variable cognitive deficits, ectodermal and musculoskeletal anomalies, and increased risk for certain malignancies. The shared mechanism of disease for this group of disorders is dysregulation of the RAS/extracellular signal-regulated kinase (ERK) and/or phosphoinositide 3-kinase/AKT signaling pathways [Tartaglia and Gelb, 2010]. Among the RASopathies, Noonan syndrome (NS; MIM #163950) is the most common and clinical variable [Roberts et al., 2013]. So far, missense germline mutations in genes encoding proteins of the RAS family of GTPases (KRAS, NRAS, RIT1, and RRAS), modulators of RAS function (PTPN11, SOS1, CBL, RASA2, and SHOC2), or downstream signal transducers (RAF1, BRAF, and MEK1), resulting in gain-of-function (GOF) effects on RAS/ERK signaling, have been reported to underlie approximately 80% of NS or related conditions [Tartaglia and Gelb, 2010; Aoki et al., 2013; Chen et al., 2014; Flex et al., 2014]. Genotype–phenotype associations have been established for several of these genes. For example, individuals with mutations in Son of Sevenless, Drosophila, homolog 1 (SOS1; MIM #182530), which encodes a member of the son-of-sevenless family of RAS guanine nucleotide exchange factors (GEFs), typically have near-normal stature and neurocognitive development but show striking ectodermal involvement [Pandit et al., 2007; Roberts et al., 2007], whereas certain mutations affecting RAF1, a serine/threonine kinase functioning as a RAS effector and initiator of the RAF/MAK/ERK cascade, and possibly those affecting RIT1, a RAS subfamily GTPase, are generally associated with early-onset hypertrophic cardiomyopathy [Pandit et al., 2007; Razzaque et al., 2007; Aoki et al., 2013].
During the last 10 years, we and others have been using a candidate gene approach, focused on genes encoding signal transducers and regulators with a role in the RAS signaling cascade to resolve the unexplained proportion of cases of NS and the other RASopathies. Since our discovery of SOS1 as a NS disease gene [Roberts et al., 2007; Tartaglia et al., 2007], Son of Sevenless, Drosophila, homolog 2 (SOS2; MIM #601247), which encodes a protein with similar function in RAS signaling, was considered an excellent candidate. Our first screening efforts, however, did not identify putative disease-causing variants, suggesting that activating mutations in this gene do not cause RASopathies or that they account for a relatively small proportion of affected subjects. To address the latter hypothesis, we initiated a multicenter collaborative screening effort using larger numbers of mutation-negative RASopathy cohorts. While this work was in progress, one paper reported the identification of heterozygous SOS2 variants in a cohort of 50 subjects with NS without an identifiable mutation in previously known disease genes [Yamamoto et al., 2015]. Among these, one was found to occur de novo in a sporadic case, and two cosegregated with the trait in two small-sized families. Here, we report that SOS2 mutations underlie a small proportion of NS cases and are associated with a clinical phenotype overlapping that resulting from SOS1 mutations. Furthermore, we demonstrate that SOS2 mutations promote enhanced activation of RAS and ERK, similar to what was observed for NS-causing SOS1 mutations [Roberts et al., 2007; Tartaglia et al., 2007]. Unlike NS-associated SOS1 mutations, however, SOS2 defects in NS are not found in multiple domains of the GEF, but specifically cluster within the Dbl homology (DH) domain.
Materials and Methods
Clinical Data and Biological Material Collection
Clinical data were obtained and biological materials were collected and stored in accordance with the ethical standards of the institutional review boards (Ospedale Pediatrico Bambino Gesù, Rome, Italy; Università Cattolica del Sacro Cuore, Rome, Italy; Istituto Mendel, Rome, Italy; Policlinico S. Orsola-Malpighi, Bologna, Italy; Hôpital Robert Debré, Paris, France; Hôpital SUD, Rennes, France; Hôpital d'Enfants, Dijon, France; Hôpital d'Enfants de la Timone, Marseille, France; University Hospital of Magdeburg, Magdeburg, Germany; Boston Children's Hospital, Boston, MA; Icahn School of Medicine at Mount Sinai, New York, NY) and after written informed consent. All subjects exhibited features fitting NS (cohort 3) or within the RASopathy phenotypic spectrum (cohorts 1 and 2), and had tested negative for mutations in previously identified RASopathy genes. The clinical diagnosis was made on the basis of standardized clinical criteria as assessed by experienced clinical geneticists. Genomic DNA was isolated from peripheral blood leukocytes and buccal mucosal epithelial cells using standard protocols. Permission was obtained to publish the photographs of subjects shown in Figure 1.
DNA Sequencing and Mutation Analysis
Mutation scanning of the entire SOS2 coding sequence (NM_006939.2; NC_000014.9 [50117128..50231878, complement]) was performed by Sanger sequencing at the US Department of Energy's Joint Genome Institute (Walnut Creek, CA), Istituto Superiore di Sanità and Ospedale Pediatrico Bambino Gesù, as previously described [Tartaglia et al., 2007].
Whole-exome sequencing (WES) was performed on 54 probands as part of the NHGRI-sponsored Centers for Mendelian Genomics program at the Yale Center for Genome Analysis (New Haven, CT) [Bamshad et al., 2012]. The WES analysis pipeline was based on the 1000 Genomes Project data analysis data pipeline, was composed from the widely used open source software projects bwa 0.7.5a [Li and Durbin, 2009], Picard 1.96, GATK 2.7 [McKenna et al., 2010; DePristo et al., 2011], snpEff 3.0 [Cingolani et al., 2012], BEDTools 2.16.2 [Quinlan and Hall, 2010], and custom-developed software, and implemented the “GATK Best Practices,” including indel realignment, deduplication, and base-quality score recalibration. Short reads were aligned to a gender- and pseudo-autosomal region-masked build of the hg19 human reference genome using bwa mem. The exome capture targets were expanded with 100-bp flanks for variant calling. Single-nucleotide variants (SNVs) and indels were called jointly with the GATK HaplotypeCaller. Variant quality score recalibration (VQSR) was used to estimate the probability that an SNV is a true variant instead of an artifact and to set the corresponding variant filter thresholds. The PASS threshold for VQSR was set to capture 99.5% of known true positives. We observed that this threshold offered a good compromise between precision and recall. Mean coverage and fractions of bases at different coverage levels were calculated with the unflanked intervals; the callable coverage of RefSeq coding exons was calculated with the flanked intervals.
The third cohort of subjects was screened by targeted resequencing. Primer pairs were designed with the IntegraGen (Evry, France) internal pipeline. Samples were amplified on an Access Array system (Fluidigm, San Francisco, CA), and equimolar pools of amplified products were sequenced on a MiSeq instrument (Illumina, San Diego, CA), using MiSeq Reagent Kit V2 cycles and paired end 2 × 150 bp. Image analysis and base calling were performed using the Real Time Analysis pipeline v. 1.14 (Illumina). Alignment of paired-end reads to the reference human genome and variant calling were carried out using the CASAVA v.1.8 pipeline (Illumina). Variant annotation was achieved using an in-house pipeline by IntegraGen. Sequencing at 25× depth covered at least 96.8% of SOS2 in all patients.
All variants identified by WES and targeted sequencing were confirmed using Sanger sequencing. When parental genomic DNAs were available, their status vis-à-vis the relevant variant was similarly assessed. When possible, paternity was confirmed by simple tandem repeat (STR) genotyping, using the AmpF/STR Identifier PCR Amplification Kit (Applied Biosystems, Waltham, MA) or the PowerPlex 16 System (Promega, Fitchburg, WI).
The frequency of the identified SOS2 variants in the general population was assessed using the Exome Aggregation Consortium's database (ExAC; http://exac.broadinstitute.org). The likelihood that variants were deleterious was evaluated using the combined annotation-dependent depletion method (CADD; http://cadd.gs.washington.edu/info) [Kircher et al., 2014], reported as scaled C-scores. A C-score threshold of 15.0 was used for declaring likely pathogenicity of missense variants. The SOS1 and SOS2 variants observed in human cancers are reported in the Catalogue of Somatic Mutations in Cancer (COSMIC; http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/).
Structural Modeling
A homology model of SOS2 (residues 6–1,043) was obtained based on the crystallographic structure of SOS1 (residues 6–1,045, pdb code 3KSY; http://www.rcsb.org/pdb/home/home.do) [Gureasko et al., 2010]. The sequence alignment was performed by using Clustal W [Larkin et al., 2007] (identity 78%) and checked manually. The structure model was built by using the Swiss Model Workspace [Arnold et al., 2006]. The software Chimera [Pettersen et al., 2004] was used for electrostatic calculations and for the structural representations reported in the figures.
Biochemical Studies
A SOS2 cDNA (Transomics, Huntsville, AL) was amplified by PCR, with the addition of a FLAG tag at the N-terminus. The amplified PCR product was subcloned into the expression vector pcDNA5/FRT/TO (Invitrogen, Carlsbad, CA). NS-associated mutations were introduced by site-directed mutagenesis.
HEK-293T cells (ATCC, Manassas, VA) were maintained in DMEM containing 10% (v/v) FBS and 100 units/ml penicillin-streptomycin (Invitrogen). Cells were seeded for 1 day before transfection with wild-type or mutant SOS2 plasmids, along with a hemagglutinin (HA)-tagged ERK expression construct at a 4:1 ratio, using FuGENE HD transfection reagent (Promega), according to the manufacturer's protocol. This design eliminates transfection efficiency as a variable, by ensuring that all cells analyzed for HA-ERK phosphorylation also express the relevant SOS2 allele. For assessing RAS activation, we generated stable transductants. Briefly, Flp-In T-REx 293 cells (Invitrogen) were cotransfected with the appropriate pcDNA5/FRT/TO expression plasmid, and the Flp recombinase-expressing plasmid pOG44 (Invitrogen) using FuGENE HD (Promega). Stable pools of transfectants were selected in hygromycin (250 μg/ml). Expression of exogenous SOS2 was induced with 1 μg/ml tetracycline 24 hr before analysis. Cells were maintained in 10% serum or serum-starved overnight, before stimulation with EGF (20 ng/ml) for various times, as indicated.
Total cell extracts were prepared in radioimmunoprecipitation assay buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 2 mM EDTA, 1% NP-40, 0.5% Na deoxycholate, 0.1% SDS), supplemented with a protease and phosphatase inhibitor cocktail (40 μg/ml phenylmethylsulfonyl fluoride, 20 mM NaF, 1 mM Na3VO4, 10 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 2 μg/ml antipapain, 2 μg/ml pepstatin A, 20 μg/ml leupeptin, and 20 μg/ml aprotinin). Lysates (20–25 μg protein) were resolved by SDS-PAGE and analyzed by immunoblotting using the following antibodies: anti-FLAG, clone M2 (Sigma–Aldrich, St. Louis, MO); anti-phospho-p44/42 MAPK and anti-phospho-MEK1/2 (Cell Signaling Technology, Beverly, MA); anti-ERK2, clone D-2 (Santa Cruz, Biotechnology, Dallas, TX); anti-ERK1/2 (Millipore, Billerica, MA). Binding of primary antibodies was detected by using IRDye infrared secondary antibodies and the Odyssey Infrared imaging system (Li-Cor Biosciences, Lincoln, NE). Quantification was performed using Odyssey V3.0 software.
RAS loading was assessed using a RAS activation assay kit (Millipore), according to manufacturer's instructions. RAS-GTP was recovered from lysates using the GST-tagged RAF-Ras binding domain of RAF1 as bait in pull-down experiments followed by SDS-PAGE and immunoblotting for RAS.
Statistical Analysis
The biochemical data for the SOS2 mutants and wild-type controls were compared using an ANOVA or a Fisher exact test, as appropriate; the threshold for declaring significance was set at P < 0.05. A Bonferroni post-hoc correction was applied to all of the ANOVA P values.
Results
SOS2 Mutation Scanning in the RASopathies
We scanned SOS2 for mutations in 150 individuals with clinically diagnosed RASopathy who had screened negative for previously discovered disease genes. We identified 19 individuals harboring nine missense variants, of which five were known polymorphisms and four were novel (Supp. Table S1). Among the latter, each observed once, three were deemed benign, as two were inherited from unaffected fathers and one was likely tolerated. The remaining variant, c.1127C>G, predicting the p.[Thr376Ser] amino acid change (Supp. Fig. S1A) had not been observed in greater than 120,000 alleles from the ExAC's database and was deemed likely deleterious using the CADD method [Kircher et al., 2014]. The variant was inherited from his mother, who exhibited features suggestive of NS. Genotyping of the mother's parents documented its de novo origin and confirmed paternity (Supp. Fig. S1A). Of note, mutation of the corresponding SOS1 residue, Thr378, had been previously reported to cause NS [Denayer et al., 2010].
Next, we scanned WES data with excellent SOS2 coverage (Supp. Fig. S1B) from another 54 mutation-negative RASopathy cases. We identified five SOS2 missense variants, including four likely polymorphisms, two recurrent and two inherited from unaffected parents. The remaining individual had a different missense nucleotide substitution affecting codon 376 (c.1126A>T, p.[Thr376Ser]), inherited from her affected mother, in whom it arose de novo (Supp. Fig. S1A). Consistent with the causative role of the SOS2 lesion in the family, no other variant affecting previously identified RASopathy genes was annotated in WES data from Subject 2.
Because the clinical features of the three affected individuals were consistent with NS (Fig. 1A; Table 1) (see below), we screened SOS2 in a third cohort, which included 61 mutation-negative subjects with clinical diagnosis of NS, using targeted resequencing. Sequencing at 25× depth covered at least 96.8% of the SOS2 coding sequence in all patients. We found a c.791C>A (p.[Thr264Lys]) and two independent c.800T>G (p.[Met267Arg]) variants (Supp. Fig. S1A) that were deemed likely pathologic due to high CADD scores, correspondence to an NS-related SOS1 mutational hotspot (p.[Thr266Lys], p.[Met269Arg], and p.[Met269Thr]), and probable GOF effects on GEF activity, based on SOS1 structural studies and expression of p.[Met269Arg] [Pandit et al., 2007; Roberts et al., 2007]. The c.791C>A allele arose de novo in that sporadic case, for whom paternity was confirmed.
| Case | 1 | 2 (mother of case 1) | 3 | 4 (daughter of case 3) | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Mutation (cDNA)a | 1127C>G | 1127C>G | 1126A>T | 1126A>T | 800T>G | 791C>A | 800T>G |
| Mutation (protein) | p.[Thr376Ser] | p.[Thr376Ser] | p.[Thr376Ser] | p.[Thr376Ser] | p.[Met267Arg] | p.[Thr264Lys] | p.[Met267Arg] |
| Sex | M | F | F | F | F | M | M |
| Age (years) | 13 | 34 | 30 | 6 | 6 | 24 | 42 |
| Perinatal findings | |||||||
| Polyhydramnios | + | na | na | + | − | na | na |
| Fetal macrosomia | − | − | + | + | + | na | na |
| Other prenatal | − | − | − | − | INT | − | − |
| Neonatal/infantile growth failure | + | na | − | − | − | − | − |
| Birth weight (kg)/birth weight SD | 2.9/−1.13 | na | na | na | 4.29/+1.89 | 3.5/−0.06 | na |
| Birth length (cm)/birth length SD | na | na | na | na | 51/+0.61 | 51/+0.31 | na |
| Birth HC (cm)/birth HC SD | na | na | na | na | 35/+0.12 | 36/+0.12 | na |
| Poor sucking | + | + | − | − | + | na | na |
| Height (cm)/height SD | 114.6/−2.53 | 161.5/−0.50 | 158/−0.82 | 113/−0.99 | 112/−0.97 | 180/+0.44 | 165/−1.65 |
| Weight (kg)/weight SD | na | n.a. | na | na | 17.9/+1.48 | na | 53/−2.03 |
| GH deficiency | + | n.a. | − | − | na | na | na |
| Craniofacial features | |||||||
| Tall forehead | + | + | + | + | − | + | + |
| Sparse eyebrows | + | + | + | + | + | + | + |
| Downslanting palpebral fissures | + | − | + | + | + | − | − |
| Hypertelorism/telecanthus | + | + | + | + | − | − | − |
| Epicanthal folds | + | + | + | + | − | − | − |
| Palpebral ptosis | + | − | + | + | + | − | + |
| Flat nasal bridge | + | − | + | + | − | − | − |
| Prominent philtrum | − | − | + | + | + | − | − |
| Thick lips | − | + | + | + | − | − | + |
| Low-set/posteriorly rotated ears | + | + | + | + | + | − | + |
| Thickened helix | + | + | + | + | + | − | − |
| Large, thick ear lobe | − | + | + | + | − | − | − |
| Cardiac anomalies | |||||||
| Pulmonary valve stenosis | − | − | − | − | − | − | + |
| Pulmonary valve dysplasia | − | − | − | − | − | − | − |
| Hypertrophic cardiomyopathy | − | − | − | − | ISH | − | + |
| Septal defects | VSD | − | ASD | − | ASD | − | − |
| Other defects | − | − | − | − | RM | − | − |
| Dermatologic findings | |||||||
| Hyperpigmented skin | − | − | − | − | − | − | − |
| Keratosis pilaris/dry skin | − | − | + | + | + | + | − |
| Sparse/absent scalp hair | + | − | + | + | − | + | na |
| Sparse/absent eyebrows | + | − | + | + | na | +b | + |
| Ulerythema ophryogenes | − | − | + | + | + | + | na |
| Curly hair | − | + | + | + | − | + | + |
| Deep palmo/plantar creases | − | − | − | − | − | − | + |
| Other features | − | − | − | − | HS | Lentiginesc | − |
| Musculoskeletal features | |||||||
| Short webbed neck | + | + | + | + | + | na | − |
| Cubitus valgus | − | − | + | + | na | na | na |
| Hyperextensible joints | − | − | + | + | − | na | + |
| Pectus deformity | + | − | + | + | +d | +d | +d |
| Central nervous system features | |||||||
| Intellectual disability | −e | − | − | −f | − | − | − |
| Brain MRI abnormalities | − | − | − | na | na | − | na |
| Seizures/EEG abnormalities | − | − | − | − | − | − | − |
| Ophthalmological anomalies | +g | − | − | − | +h | − | na |
| Gastrointestinal anomalies | − | +i | − | − | na | − | − |
| Cryptorchidism | + | + | + | ||||
| Urogenital anomalies | − | − | − | na | +j | ||
| Hematologic anomalies | − | − | − | − | na | +k | − |
| Lymphatic anomalies | − | + | − | − | +l | +m | − |
| Miscellaneous | − | − | − | − | +n | BIH | +o |
- a Position referred to the A of the ATG translation initiation codon in the reference cDNA sequence (NM_006939.2).
- b Normalized by 24 years.
- c Multiple lentigines spread on the face and neck.
- d Excavatum.
- e Mild learning difficulties.
- f IQ 120.
- g Hyperopia.
- h Right: posterior embryontoxin; left: enlarged corneal nerves.
- i Gastric malrotation.
- j Bilateral uretero-pelvic junction stenosis.
- k Easy bruising.
- l Marked lymphedema of the right leg (around 6 years).
- m Lymphedema of hands and feet (onset at 15 years).
- n Superinfection of right buttock cyst.
- o Charcot-Marie-Tooth disease type 2, onset in infancy (unidentified gene), hypereosinphilia.
- BIH, bilateral inguinal hernia; HC, head circumference; HS, hyperelastic skin; INT, increased nuchal translucency; ISH, isolated septal hypertrophy; NA, data not available; RM, rhabdomyoma of the right ventricle (spontaneously resolved); SD, standard deviation (based on CDC growth charts 2000).
All SOS2 variants described in this report have been submitted to the NSEuronet mutation database (https://nseuronet.com/php/index.php).
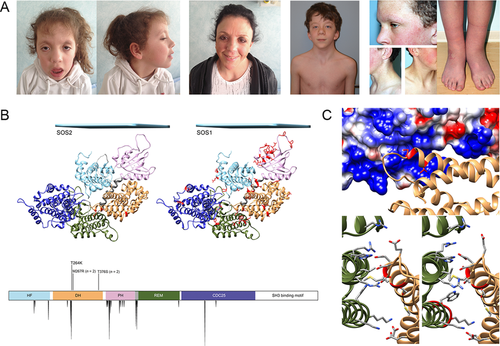
Molecular Modeling of NS-Associated SOS2 Mutations
Similar to the structurally and functionally related SOS1, SOS2 stimulates the release of GDP from RAS, promoting the conversion of the GTPase from the inactive, GDP-bound to the active, GTP-bound form [Nimnual and Bar-Sagi, 2002]. SOS2 is a large multidomain protein characterized by an N-terminal regulatory portion including two tandemly arranged histone-like folds, which are followed by DH domain and a pleckstrin-homology (PH) domain, and a C-terminal catalytic region comprising the RAS exchanger motif (REM) and CDC25 domains, followed by a tail containing docking sites for adaptor proteins required for receptor anchoring (Fig. 1B). In SOS1, GEF activity is controlled principally by two binding sites for RAS: the catalytic site, which is located within the CDC25 domain, and a distal site involving two adjacent regions of the CDC25 and REM domains. The latter domain positively modulates GEF activity by promoting a conformational change at the active site that allows GDP-RAS to bind [Margarit et al., 2003]. The majority of SOS1 mutations causing NS affect residues that are implicated in the maintenance of SOS1 in its autoinhibited conformation [Roberts et al., 2007; Tartaglia et al., 2007; Lepri et al., 2011]. Among these, a class of mutations involves residues that participate in the autoinhibitory interaction of the DH and REM domains that blocks RAS access to the allosteric site. These mutations directly affect the stability of the inactive conformation of the GEF directly by disrupting the inhibitory interdomain bonding network at the distal site.
To analyze the functional impact of the three disease-associated missense variants, all altering conserved residues in the DH domain (Fig. 1B), a model of autoinhibited SOS2 (residues 6–1,043) was obtained using the crystallographic structure of SOS1 (residues 6–1,045, pdb code 3KSY) as the template [Gureasko et al., 2010]. Based on this model, the p.[Thr264Lys] and p.[Met267Arg] substitutions insert positively charged residues in the anionic DH region that interfaces with a cationic region of the REM domain, thereby reducing electrostatic attraction (Fig. 1C). Similar to what was observed for SOS1 mutations affecting certain residues (p.[Thr266Lys], p.[Met269Arg/Thr], p.[Lys728Ile], p.[Trp729Leu], and p.[Ile733Phe]), these substitutions were predicted to activate SOS2 by destabilizing the DH–REM interaction, allowing RAS to access the allosteric site [Sondermann et al., 2004]. Thr376 is solvent-exposed, so the effect of the p.[Thr376Ser] substitution was unclear.
Functional Effects of NS-Associated SOS2 Mutations
To test the GOF role of NS-associated SOS2 mutations predicted by our molecular modeling analysis, the effects exerted by these mutations on RAS-ERK signaling were assessed by comparing the levels of ERK and MEK phosphorylation as well as that of GTP-bound RAS in transient transfection assays. Consistent with the predicted activating role of the three amino acid substitutions, expression of the SOS2Thr264Lys, SOS2Met267Arg, and SOS2Thr376Ser mutants promoted enhanced phosphorylation of both endogenous and exogenous ERK compared with that observed in cells expressing the wild-type protein (Fig. 2A). Similarly, enhanced activation of endogenous MEK was observed in cells expressing these SOS2 mutants. Of note, the SOS2 mutants engendered different degrees of hyperactivation. ERK and MEK were constitutively active in SOS2Thr264Lys- and SOS2Met267Arg-expressing cells, whether randomly growing or serum-starved. By contrast, cells expressing the SOS2Thr376Ser mutant exhibited enhanced and protracted activation of these kinases, but activation remained dependent on growth factor stimulation. The increased GEF activity of the NS-associated SOS2 mutants is consistent with the structural analyses. Moreover, our biochemical analyses indicate a differential impact of NS-associated mutations at the level of SOS2 activation.
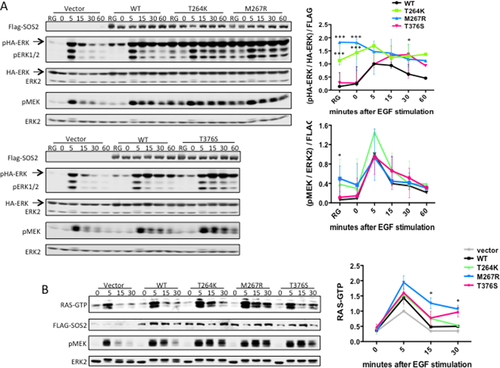
We also assessed RAS activation by generating stable transductants in Flp-In T-Rex 293 cells, which allow tetracycline-inducible expression of SOS2 constructs at near endogenous levels. Compared with the effects of wild-type SOS2, expression of all three SOS2 mutants led to increased RAS activation, shown as higher levels of GTP-bound RAS. These findings are consistent with a direct GOF effect on the RAS-GEF activity of NS-associated SOS2 mutations.
Overall, these data provide evidence that, similar to what has been reported previously for NS-causative SOS1 mutations, NS-associated SOS2 mutations promote enhanced activation of RAF-MEK-ERK signaling cascade, an effect that is directly mediated by their enhanced RAS-GEF activity.
Clinical Features Observed in NS due to SOS2 Mutations
Detailed clinical data were collected for the six affected individuals with the four SOS2 missense mutations to explore possible genotype–phenotype correlations. All subjects displayed typical NS facial features (Fig. 1A). Notably, height was normal in all but one individual, and all individuals exhibited normal or nearly normal neurocognitive status at 6–42 years (Table 1). By contrast, ectodermal involvement was striking and similar to that seen in SOS1 mutation-associated NS patients, including facial keratosis pilaris, sparse scalp hair, and ulerythema ophryogenes (Fig. 1A; Table 1). Cardiac involvement was typical for NS. Overall, the genotype–phenotype associations observed with the SOS2 mutations resembled those established for SOS1 mutations.
Discussion
Here, we report that SOS2 is mutated in NS, confirming the recent report of a Brazilian cohort from Yamamoto et al. (2015). We provide the first data indicating that disease-causing SOS2 mutations promote enhanced GEF function in SOS2, resulting in enhanced signaling through RAS and the RAF-MEK-ERK cascade. Finally, the available clinical records support the idea that SOS2 mutations are associated with a phenotype resembling that previously associated with SOS1 mutations.
Mutations in SOS1 constitute approximately 10% of NS cases. In contrast, SOS2 mutations are estimated to account for a far smaller percentage of cases in the three cohorts we analyzed. Remarkably, all of the eight independent SOS2 mutations underlying NS were found to alter only one of three amino acids (i.e., Thr264, Met267, and Thr376), all located in the DH domain. This is strikingly different from NS-associated SOS1 mutations that arise at multiple hotspots (Fig. 1B) [Pandit et al., 2007; Roberts et al., 2007; Lepri et al., 2011]. The two SOS proteins are nearly 70% homologous, expressed ubiquitously, and activate RAS proteins. SOS2, however, has weaker biological effects. For example, Sos1 deficiency in mice is embryonic lethal, whereas mice without Sos2 are normal [Esteban et al., 2000; Qian et al., 2000]. SOS2 has been shown to be much less stable than SOS1 because of its accelerated degradation via a ubiquitin-dependent process [Nielsen et al., 1997]. Furthermore, expression of myristoylated SOS1 promotes cell transformation, whereas similarly tagged SOS2 cannot. Consistent with this idea, several germline NS-causing SOS1 GOF mutations at three major hotspots within the DH and PH domains as well as the helical linker occur as sporadic mutations in cancers (as reported in COSMIC), whereas cancer-associated SOS2 mutations only affect the DH domain. The driving force behind the SOS2 DH domain “hotspot” shared with SOS1 is likely biochemical, not genetic. Both mutants are constitutively active; neither mutation arose at a CpG dinucleotide and both are transversions. Although the precise mechanism is unclear, the same is likely true of the two nucleotide changes resulting in the p.[Thr376Ser] substitution, which also did not arise at CpG dinucleotides, are transversions and for which a comparable SOS1 substitution has been observed once in cancer.
Our data, which represent the first biochemical characterization of NS-causing SOS2 mutations, suggest that different mechanisms likely drive the functional dysregulation of SOS2 and, in turn, aberrant RAS activation. Mutations affecting Thr264 and Met267, which are located in the surface of the DH domain that mediates the autoinhibitory interaction with the REM domain, were shown to constitutively upregulate MEK and ERK activity. The corresponding residues in SOS1, Thr266 and Met269, are among the most common site of mutations in NS. In particular, Met269, which is mutated in approximately 10% of cases bearing SOS1 mutations, interacts directly with residues of the REM domain implicated in RAS binding [Sondermann et al., 2004]. As previously documented for SOS1, these mutations are predicted to affect the stability of the inactive conformation of the protein directly by disrupting the inhibitory interdomain bonding network at the distal site [Lepri et al., 2011]. On the other hand, a different pattern of signaling dysregulation was observed in cells overexpressing SOS2Thr376Ser. In these cells, ERK activation was enhanced and protracted but retained dependence on EGF stimulation. While the solvent-exposed position of Thr376 suggests a possible role in the interaction of the GEF with signaling partners and/or modulators, the available structural data do not allow us to recognize any functional clue for this recurrent NS-causing SOS2 substitution.
Analysis of the clinical data documented that mutations in SOS2 are associated with a consistent phenotype that unambiguously falls within the NS clinical spectrum, but is apparently characterized by a high prevalence of ectodermal features and low occurrence of short stature and cognitive impairment compared with what is observed in the NS general population. Overall, the SOS2-associated NS phenotype closely resembles that associated with SOS1 mutations. Notably, both groups of patients enjoy excellent neurodevelopment, which is likely attributable to the developmental stage-dependent expression of RAS GEFs. While SOS proteins activate RAS downstream from N-methyl-D-aspartate glutamate receptors in cortical neurons of the developing and neonatal central nervous systems (CNS), there is a switch to RAS guanine nucleotide-releasing factors 1 and 2 later in life [Tian et al., 2004]. Consistent with evidence that NS-associated PTPN11 mutations perturb CNS homeostasis, not brain development [Pagani et al., 2009; Lee et al., 2014], restoration of normal CNS RAS signaling in children with SOS mutations enables normal development. Hence, our findings provide further hope that therapies reducing RAS signaling in children with NS-causing mutations in non-SOS genes would improve their developmental trajectories.
Overall, the present work provides evidence that a narrow spectrum of activating missense mutations in SOS2 account for a small proportion of NS, and that subjects heterozygous for a germline SOS2 mutation exhibit a distinctive phenotype resembling that of SOS1 mutations, providing new clinically valuable information for diagnosis and more effective patient management.
Acknowledgments
We thank the families who participated in the study, Serenella Venanzi (Istituto Superiore di Sanità, Rome, Italy), Natacha Fillot and Nathalie Pouvreau (Robert Debré Hospital, Paris, France) for technical assistance, and Guillaume Gauchotte (Nancy University Hospital, Nancy, France) for the tissue biopsy specimen (father of patient 6).
Several authors (B.D.G., B.G.N., A.E.R., and M.T.) are inventors for patents that have been licensed by their institutions for gene testing for NS and for which they receive royalties. By virtue of expanding the utility of testing, the work described in this manuscript could theoretically increase those royalties.
B.G.N. is a Canada Research Chair, Tier 1. L.A.P.’s research was conducted at the E.O. Lawrence Berkeley National Laboratory, performed under Department of Energy Contract DE-AC02-05CH11231.




