Morquio A Syndrome-Associated Mutations: A Review of Alterations in the GALNS Gene and a New Locus-Specific Database
Communicated by Hans R. Waterham
ABSTRACT
Morquio A syndrome (mucopolysaccharidosis IVA) is an autosomal recessive disorder that results from deficient activity of the enzyme N-acetylgalactosamine-6-sulfatase (GALNS) due to alterations in the GALNS gene, which causes major skeletal and connective tissue abnormalities and effects on multiple organ systems. The GALNS alterations associated with Morquio A are numerous and heterogeneous, and new alterations are continuously identified. To aid detection and interpretation of GALNS alterations, from previously published research, we provide a comprehensive and up-to-date listing of 277 unique GALNS alterations associated with Morquio A identified from 1,091 published GALNS alleles. In agreement with previous findings, most reported GALNS alterations are missense changes and even the most frequent alterations are relatively uncommon. We found that 48% of patients are assessed as homozygous for a GALNS alteration, 39% are assessed as heterozygous for two identified GALNS alterations, and in 13% of patients only one GALNS alteration is detected. We report here the creation of a locus-specific database for the GALNS gene (http://galns.mutdb.org/) that catalogs all reported alterations in GALNS to date. We highlight the challenges both in alteration detection and genotype–phenotype interpretation caused in part by the heterogeneity of GALNS alterations and provide recommendations for molecular testing of GALNS.
Introduction
Morquio A syndrome (mucopolysaccharidosis IVA, MPS IVA; MIM #253000) is an autosomal recessive lysosomal storage disorder caused by deficient activity of the enzyme N-acetylgalactosamine-6-sulfatase (GALNS; also known as N-acetylgalactosamine-6-sulfate sulfatase or galactosamine (N-acetyl)-6-sulfate sulfatase; MIM #612222) due to mutations in the GALNS gene. Deficient GALNS activity causes accumulation of the glycosaminoglycans (GAGs) keratan sulfate (KS) and chondroitin-6-sulfate (C6S) in multiple tissues, which leads to skeletal and connective tissue abnormalities together with pulmonary limitations and cardiac pathology [Harmatz et al., 2013; Montaño et al., 2007; Yasuda et al., 2013]. Morquio A is a rare disorder, with an estimated incidence ranging from one in 76,000 births in Northern Ireland to one in 640,000 births in Western Australia [Hendriksz et al., 2013a; Nelson, 1997; Nelson et al., 2003]. The clinical presentation of Morquio A is characterized by variable expressivity, but even patients with more attenuated forms of Morquio A are significantly affected and suffer substantial disease burden [Montaño et al., 2007; Montaño et al., 2008; Tomatsu et al., 2011; Hendriksz et al., 2013a; Harmatz et al., 2013].
Clinical suspicion of Morquio A syndrome typically leads to screening urine GAG analysis or screening enzyme activities in dried blood spot samples, but the gold standard for diagnosis of Morquio A is demonstrating deficient GALNS enzyme activity in either leukocytes or fibroblasts together with wild-type activity of appropriate control enzymes to confirm sample integrity and exclude other disorders that can reduce GALNS enzyme activity, such as multiple sulfatase deficiency or mucolipidosis type II/III [Wood et al., 2013]. Molecular analysis can confirm the Morquio A diagnosis and aid genetic counseling by detecting the causative alterations in the GALNS gene [Tylee et al., 2013; Wood et al., 2013].
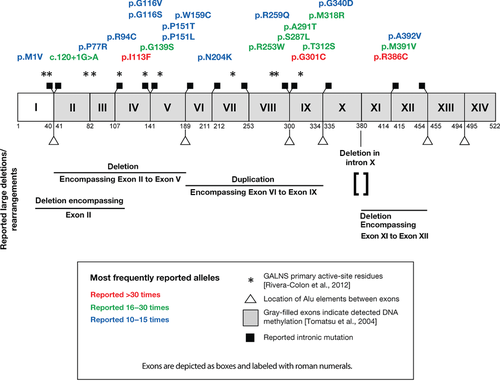
The GALNS gene is approximately 50 kb in length and contains 14 exons, producing a 2,339 base pair mRNA that encodes a 522-amino acid protein (Fig. 1) [Tomatsu et al., 1991; Nakashima et al., 1994]. The alterations in the GALNS gene associated with Morquio A are numerous and heterogeneous and have been found throughout the coding sequence and flanking splice sites; as of 2005, 148 unique gene alterations were known [Tomatsu et al., 2005b] and many more have since been reported. The most common GALNS gene alterations are missense alterations [Tomatsu et al., 2005b; Hendriksz et al., 2013a]. DNA methylation at CpG sites has been detected in every exon but the first, and the inappropriate repair of spontaneous deamination events is thought to lead to transition mutations at these sites [Tomatsu et al., 2004]. Multiple introns contain Alu repetitive elements (http://genome.ucsc.edu/, February 2009 Assembly)[Meyer et al., 2013], which can undergo recombination and potentially lead to large deletions and/or rearrangements. Even the most commonly detected GALNS alleles are relatively uncommon, although some alleles are far more prevalent in certain regions or ethnic group subpopulations, possibly due to founder effects in which one or a few GALNS gene alterations were present in populations started by a small number of individuals [Tomatsu et al., 2005b; Wood et al., 2013; Yamada et al., 1998; Kato et al., 1997]. This heterogeneity can pose challenges for the interpretation of molecular testing results from patients with Morquio A because detection of novel GALNS gene alterations of unknown significance may occur relatively frequently.
Methods
We performed a literature search for GALNS gene alterations reported from patients with Morquio A when at least one altered GALNS allele was reported. GALNS gene alteration data were gathered from the primary reports. When the information was available, the gene alterations were recorded as patient genotypes (i.e., the two GALNS alleles reported from a patient); however, there were 81 instances where GALNS gene alterations were reported in tabular lists without information about their occurrence in patients. Some older reports of GALNS gene alterations were adjusted so that all DNA and protein sequence numbering is based on the GALNS cDNA sequence (GenBank entry NM_000512.4), and the DNA sequence position “1” corresponds to the A of the initial ATG in the reference sequence. In some cases, errors in original publications have been corrected based on subsequent reports or author communications (Supp. Table S1). Sequence variant descriptions follow recommendations of the Human Genome Variation Society [den Dunnen and Antonarakis, 2000]. Assessed patient GALNS genotypes, assessments of disease state, and patient geographic/ethnic affiliations are as reported in the referred original publications(s) of that gene alteration.
Reported GALNS gene alterations associated with Morquio A were classified by mutation type, in many cases reporting deduced changes to the GALNS amino acid sequence: missense alterations, including alterations in the initial GALNS ATG and nucleotide substitutions eliminating the GALNS stop codon; nonsense alterations; “insertion and/or deletion” alterations; and “intronic” alterations, nucleotide substitutions in an intron (insertion/deletions occurring in introns are classified as “insertion and/or deletion”).
Results and Discussion
Alterations in the GALNS Gene from Patients with Morquio A
The information presented here serves as an update and augmentation to a previous comprehensive summary of alterations in the GALNS gene associated with Morquio A syndrome [Tomatsu et al., 2005b]. In addition to the inclusion of patient data and gene alterations published since 2005, the data were gathered in the context of patient genotypes (i.e., the two GALNS alleles reported from a patient) when available because this allows additional analyses not possible previously. Additionally, three new genotypes from patients with Morquio A were recently published [Lachman et al., 2014], including a patient with the novel GALNS gene alterations c.317A.G (p.Asn106Ser) and c.553delG (p.Glu185Argfs14). These data have been added to Supp. Table S2 only and are not included in this manuscript's analyses. This Mutation Update is a retrospective analysis of alterations in the GALNS gene as reported in the primary literature. However, the possibility exists that a GALNS alteration reported to be associated with Morquio A disease may not be causative, and so caution is warranted in the interpretation of rare GALNS alterations with unclear molecular consequences.
Genotypes from Patients with Morquio A
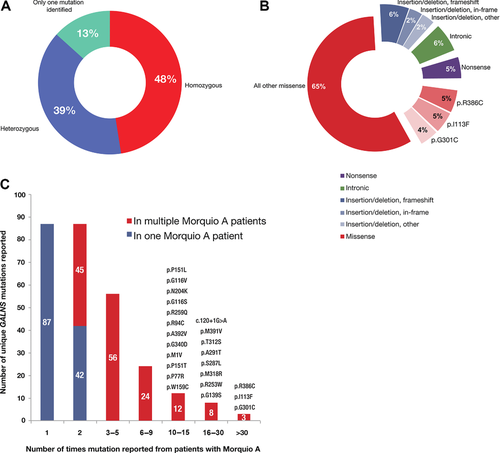
In a retrospective analysis, we identified from the literature 541 published genotypes of patients with Morquio A syndrome for which at least one alteration in the GALNS gene was reported (Supp. Table S2). Of these 541 patients, 257 patients (48%) were assessed as being homozygous for an identified GALNS gene alteration, 212 patients (39%) were assessed as being heterozygous for two identified gene alterations, and 72 patients (13%) had an alteration in only a single GALNS allele identified (Fig. 2A). Because parental genotyping (to confirm a patient's assessed genotype) and/or analysis for large deletions and other rearrangements are often not performed, an unknown subset of the patients assessed as homozygous actually may be heterozygous for the detected GALNS gene alteration and an undetected gene alteration (see Clinical and Diagnostic Strategies).
GALNS Alteration Types and Frequencies
First, we examined distinct unique gene alterations in GALNS regardless of how frequently they are reported, counting multiple reports of the same gene alteration only once. The 541 patients with Morquio A from whom information about GALNS homozygosity or heterozygosity was reported and the 81 instances of GALNS alleles reported in tabular form (where homozygosity or heterozygosity was not reported) together contain 277 distinct reported gene alterations. Counting multiple reports of the same mutation as one distinct gene alteration, missense alterations are the most frequently reported distinct alterations in GALNS from patients with Morquio A: distinct missense alterations account for 185 (67%) of the 277 reported distinct gene alterations, in close agreement with other published results [Tomatsu et al., 2005b; Hendriksz et al., 2013a]. The next most common detected gene alteration type is insertions and/or deletions (47 out of 277 distinct gene alterations; 17%). Of the 47 detected distinct insertions and/or deletions, 28 are predicted to cause frameshifts, 11 are in-frame deletions or insertions, and eight likely have more complex effects. Detected deletions, both small and large, outnumber detected insertions 39 to 8. Few large deletions or complex rearrangements have been reported to date, possibly due to under-detection (see Clinical and Diagnostic Strategies). Intronic changes (excluding deletions and/or insertions, which are considered above) account for 24 of the 277 distinct gene alterations (9%); 22 of these gene alterations affect one of the −1, −2, +1, or +2 nucleotides. Nonsense changes account for 21 of the 277 distinct gene alterations (8%) in GALNS reported from patients with Morquio A.
Next, we examined the frequency at which alterations in the GALNS gene were reported from patients with Morquio A and totaled all reports of the same GALNS allele (Supp. Table S3, Fig. 2B). This is different from the above analysis of gene alterations types as a function of the 277 distinct gene alterations. This analysis is performed with a different denominator unit, the number of GALNS alleles. The 541 patients with Morquio A have 1,010 reported mutant GALNS alleles and 72 instances in which the second mutant GALNS allele was not identified. The 1010 reported GALNS alleles from patient GALNS genotypes were combined with 81 alleles reported in tabular form to yield 1,091 reported mutant GALNS alleles in patients with Morquio A. Of the 1,091 detected alleles, 858 are missense alleles (79% of all alleles; Fig. 2B). Again, this analysis differs from that of the previous paragraph: from patients with Morquio A, missense mutations are both the most frequently reported distinct alterations in the GALNS gene (67% of 277 distinct alterations) and the most frequently reported alleles (79% of 1,091 detected GALNS alleles). Of the 1,091 mutant GALNS alleles reported from patients, the three most commonly reported alleles together represent only 152 of 1,091 reported alleles (14%; Fig. 2B), again demonstrating the heterogeneity of alterations in the GALNS gene from patients with Morquio A [Tomatsu et al., 2005b](Fig. 2C). The allele most frequently reported is the missense change c.1156C>T (p.Arg386Cys), reported 55 times (55 of 1,091; 5%); the second most frequently reported allele was the missense change c.337A>T (p.Ile113Phe), reported 52 times (52 of 1,091; 5%); and the third most reported allele was the missense change c.901G>T (p.Gly301Cys), reported 45 times (45 of 1,091; 4%). These same three alleles are the most frequent both in this data set and in Tomatsu et al. (2005b), but their relative frequency is reduced in this data set. Compared with Tomatsu et al. (2005b), here c.1156C>T (p.Arg386Cys) is 5% of reported GALNS alleles but 8.9% previously; c.337A>T (p.Ile113Phe) is 5% of reported alleles here but 5.7% previously; and c.901G>T (p.Gly301Cys) is 4% of reported alleles here but 6.8% previously.
Notably, many GALNS alleles have only been reported in a single patient: 42 GALNS gene alterations have only been reported in the homozygous state from a single patient and 86 of 277 gene alterations (31%) have only been reported once (Fig. 2C).
GALNS Alleles and Geography
Worldwide, the ten most frequently reported GALNS gene alterations from patients with Morquio A (Table 1) are c.1156C>T (p.Arg386Cys; reported 55 times), c.337A>T (p.Ile113Phe; reported 52 times), c.901G>T (p.Gly301Cys; reported 45 times), c.120+1G>A (reported 23 times), c.1171A>G (p.Met391Val; reported 22 times), c.935C>G (p.Thr312Ser; reported 22 times), c.871G>A (p.Ala291Thr; reported 20 times), c.860C>T (p.Ser287Leu; reported 20 times), c.953T>G (p.Met318Arg; reported 19 times), and c.757C>T (p.Arg253Trp; reported 18 times).
| Allele | Number detected | Percentage of that allele's total | Percentage of all detected alleles |
|---|---|---|---|
| p.Arg386Cys | 55 | 100 | 5.0 |
| Spanish | 9 | 16 | 0.8 |
| Argentine | 6 | 11 | 0.5 |
| Chinese | 5 | 9 | 0.5 |
| Italian | 4 | 7 | 0.4 |
| Colombian | 3 | 5 | 0.3 |
| Polish | 3 | 5 | 0.3 |
| Turkish | 3 | 5 | 0.3 |
| Chilean | 3 | 5 | 0.3 |
| All other countries/ethnicities | 14 | 35 | 1.3 |
| p.Ile113Phe | 52 | 100 | 4.8 |
| Irish | 27 | 52 | 2.5 |
| British | 15 | 29 | 1.4 |
| British/Irish | 3 | 6 | 0.3 |
| All other countries/ethnicities | 7 | 13 | 0.6 |
| p.Gly301Cys | 45 | 100 | 4.1 |
| Colombian | 16 | 36 | 1.5 |
| Portuguese | 6 | 13 | 0.5 |
| Spanish | 5 | 11 | 0.5 |
| All other countries/ethnicities | 10 | 40 | 0.9 |
| c.120+1G>A | 22 | 100 | 2.0 |
| Tunisian | 20 | 91 | 1.8 |
| All other countries/ethnicities | 2 | 9 | 0.2 |
| p.Thr312Ser | 22 | 100 | 2.0 |
| Irish | 14 | 64 | 1.3 |
| British/Irish | 3 | 14 | 0.3 |
| All other countries/ethnicities | 5 | 23 | 0.5 |
| p.Met391Val | 22 | 100 | 2.0 |
| French Canadian | 7 | 32 | 0.6 |
| French | 4 | 18 | 0.4 |
| Canadian Caucasian | 3 | 14 | 0.3 |
| American caucasian, German | 2 | 9 | 0.2 |
| All other countries/ethnicities | 6 | 27 | 0.5 |
| p.Ala291Thr | 20 | 100 | 1.8 |
| Asian-multiethnic | 8 | 40 | 0.7 |
| British | 4 | 20 | 0.4 |
| Finnish | 3 | 15 | 0.3 |
| Pakistani | 2 | 10 | 0.2 |
| Chinese | 1 | 5 | 0.1 |
| Japanese | 1 | 5 | 0.1 |
| All other countries/ethnicities | 1 | 5 | 0.1 |
| p.Ser287Leu | 20 | 100 | 1.8 |
| Middle Eastern | 4 | 20 | 0.4 |
| Turkish | 3 | 15 | 0.3 |
| Spanish | 2 | 10 | 0.2 |
| Polish | 2 | 10 | 0.2 |
| Greek | 2 | 10 | 0.2 |
| Macedonian | 2 | 10 | 0.2 |
| Austrian | 1 | 5 | 0.1 |
| New Zealander | 1 | 5 | 0.1 |
| Irish/Italian/Polish | 1 | 5 | 0.1 |
| All other countries/ethnicities | 2 | 10 | 0.2 |
| p.Met318Arg | 19 | 100 | 1.7 |
| Chinese | 11 | 58 | 1.0 |
| Taiwanese | 3 | 16 | 0.3 |
| Other | 2 | 11 | 0.2 |
| South-East Asian | 2 | 11 | 0.2 |
| Japanese | 1 | 5 | 0.1 |
| p.Arg253Trp | 18 | 100 | 1.6 |
| Pakistani | 16 | 89 | 1.5 |
| All other countries/ethnicities | 2 | 11 | 0.2 |
- “All other countries/ethnicities” includes not given.
In addition to overall differences in the frequency that individual GALNS alleles are reported, there are differences in reported allele frequencies in specific subpopulations, which makes some alleles more or less common than might be expected. This analysis relied on information of patients’ countries, regions, or ethnicities as provided by the references, which may not have been collected in a consistent manner. However, it is relevant to consider both the reported country/regional, and/or ethnic affiliations associated with patients with Morquio A, because in aggregate they reveal substantial differences in the frequency of different GALNS alleles in different populations. For example, the splice site mutation c.120+1G>A is the fourth most frequently reported GALNS gene alteration from patients with Morquio A, but most reports are for patients from Tunisia (Table 1). The missense alteration c.337A>T (p.Ile113Phe) is the second most frequently reported GALNS gene alteration from patients with Morquio A, but it is most frequently reported in patients from the British Isles (Table 1), and for Morquio A patients identified as of “Irish” descent, c.337A>T (p.Ile113Phe) represents 43% of all reported GALNS gene alterations (Table 2). Other localities beyond those listed in Table 2 also have high reported rates of individual alterations; for example, the gene alteration c.901G>T (p.Gly301Cys) is 50% of all reported alleles from Colombian patients with Morquio A. The high frequency of a specific GALNS gene alteration in some populations could be due to founder effects.
| Reported country/ethnicity | Alleles detected (all alleles, including unidentified) | Percentage of that country's total | Percentage of all detected alleles |
|---|---|---|---|
| Chinese | 93 (118) | 100 | 8.5 |
| p.Met318Arg | 11 | 12 | 1.0 |
| p.Gly340Asn | 9 | 10 | 0.8 |
| p.Leu366Pro | 7 | 8 | 0.6 |
| All other alleles | 66 | 71 | 6.0 |
| Japanese | 52 (67) | 100 | 4.8 |
| p.Asn204Lys | 8 | 15 | 0.7 |
| Double gene deletion | 8 | 15 | 0.7 |
| p.His430GlnfsTer71 | 5 | 10 | 0.5 |
| Double deletion | 5 | 1 | 0.5 |
| c.121–2A>G | 4 | 8 | 0.4 |
| All other alleles | 22 | 42 | 2.0 |
| Irish | 60 (63) | 100 | 5.5 |
| p.Ile113Phe | 27 | 45 | 2.5 |
| p.Thr312Ser | 14 | 23 | 1.3 |
| All other alleles | 19 | 32 | 1.7 |
| British | 52 (62) | 100 | 4.8 |
| p.Ile113Phe | 15 | 29 | 1.4 |
| p.Ala291Thr | 4 | 8 | 0.4 |
| All other alleles | 33 | 63 | 3.0 |
| Asian-multiethnic | 58 (58) | 100 | 5.3 |
| p.Gly116Val | 12 | 21 | 1.1 |
| p.Ala291Thr | 8 | 14 | 0.7 |
| p.Pro151Leu | 7 | 12 | 0.6 |
| p.Ala392Val | 6 | 10 | 0.5 |
| p.Leu36Arg | 6 | 10 | 0.5 |
| p.Gly139Ser | 4 | 7 | 0.4 |
| All other alleles | 15 | 26 | 1.4 |
| Turkish | 49 (51) | 100 | 4.7 |
| p.Glu112ArgfsTer15 | 8 | 1 | 0.7 |
| p.Met494Val | 4 | 8 | 0.4 |
| p.Trp141Arg | 4 | 8 | 0.4 |
| p.Leu390Ter | 4 | 8 | 0.4 |
| p.His236ArgfsTer25 | 4 | 8 | 0.4 |
| p.Arg386Cys | 3 | 6 | 0.3 |
| p.Ser287Leu | 3 | 6 | 0.3 |
| All other alleles | 19 | 39 | 1.9 |
| Middle Eastern | 47 (48) | 100 | 4.4 |
| p.Arg94Cys | 6 | 13 | 0.5 |
| p.Gly201Glu | 6 | 13 | 0.5 |
| p.Ser287Leu | 4 | 9 | 0.4 |
| p.Trp159Cys | 4 | 9 | 0.4 |
| p.Asp233Asn | 4 | 9 | 0.4 |
| c.121-1G>A | 4 | 9 | 0.4 |
| All other alleles | 19 | 40 | 1.8 |
| Italian | 45 (45) | 100 | 4.1 |
| p.Arg386Cys | 4 | 9 | 0.4 |
| p.Met1Val | 8 | 18 | 0.7 |
| p.Trp10Ter | 4 | 9 | 0.4 |
| p.Phe167Val | 4 | 9 | 0.4 |
| p.Pro125Leu | 3 | 7 | 0.3 |
| p.Ala107Thr | 3 | 7 | 0.3 |
| All other alleles | 19 | 42 | 1.7 |
| Brazilian | 37 (38) | 100 | 3.5 |
| p.Ser341Arg | 5 | 14 | 0.5 |
| p.Gly116Ser | 4 | 11 | 0.4 |
| p.Val239Phe | 4 | 11 | 0.4 |
| p.Cys165Tyr | 4 | 11 | 0.4 |
| p.Asn164Thr | 3 | 8 | 0.3 |
| p.Ala203Val | 3 | 8 | 0.3 |
| p.Arg386Cys | 2 | 5 | 0.2 |
| p.Gly301Cys | 2 | 5 | 0.2 |
| p.Arg94Cys | 2 | 5 | 0.2 |
| p.Val16Glu | 2 | 5 | 0.2 |
| p.Leu307Pro | 2 | 5 | 0.2 |
| p.Glu51Lys | 2 | 5 | 0.2 |
| All other alleles | 2 | 5 | 0.3 |
| Spanish | 36 (36) | 100 | 3.3 |
| p.Arg386Cys | 9 | 25 | 0.8 |
| p.Gly301Cys | 5 | 14 | 0.5 |
| Exon 5 skipping | 5 | 14 | 0.5 |
| p.Tyr254Cys | 5 | 14 | 0.5 |
| p.Ser287Leu | 2 | 6 | 0.2 |
| p.Gly139Ser | 2 | 6 | 0.2 |
| All other alleles | 8 | 22 | 0.7 |
Genotype–Phenotype Correlation
Morquio A disease severity descriptions historically focused primarily on height and growth, which measure only a specific manifestation of a disease that affects multiple body systems with variable expressivity [Harmatz et al., 2013; Montaño et al., 2007; Yasuda et al., 2013]. Patients have physical limitations resulting from the skeletal, connective tissue, respiratory, and cardiac manifestations of Morquio A; endurance, mobility, and the ability to complete everyday activities (e.g., clipping nails or brushing hair) are also relevant measures of disease severity [Harmatz et al., 2013]. The International Morquio A Registry documented the natural course of Morquio A disease and showed that frequent surgical interventions are required by patients with Morquio A [Montaño et al., 2007]. Clinical experience demonstrates that the attenuated nature of the disease is subjective and likely underestimated when only height/growth are considered as a measure of disease severity. A pair of siblings with Morquio A seen in clinical practice illustrates this: one sibling was very short but ambulatory and with minimal medical problems; whereas, the other sibling was taller but could not walk independently and had a history of surgeries related to Morquio A (personal communication, P. Harmatz). As de novo GALNS mutations are rare and these sibs are expected to have the same gene alterations, other genetic factors and their individual medical histories may also contribute to these differences. These siblings illustrate that height or growth-based metrics alone may not capture fully the effect of Morquio A on a patient.
The genotype–phenotype relationship of 148 Morquio A GALNS gene alterations was previously assessed [Tomatsu et al., 2005b]: (1) by the homozygosity of the gene alteration, (2) by the level of residual activity of the GALNS enzyme based on in vitro expression study, (3) based on predictions of the effect of the gene alteration on the protein structure, and (4) in patients with attenuated phenotypes with an allele permitting residual enzyme activity, which would be dominant over an allele permitting no activity. Tomatsu et al. [2005b] found 31 gene alterations associated with an attenuated phenotype and 101 gene alterations associated with a severe phenotype but also noted the challenges in associating GALNS genotypes with Morquio A phenotypes. Rivera-Colon et al. (2012) also recently summarized GALNS gene alterations with associated disease severity, which, in the existing Morquio A literature, generally refers to a growth rate phenotype.
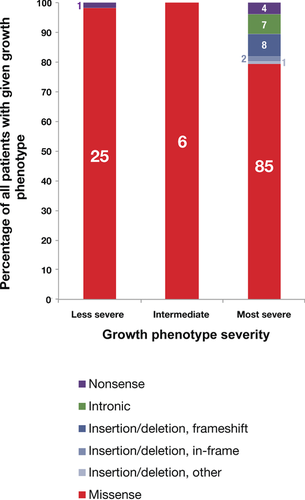
In a retrospective analysis, we summarize the mutation types found in patients with Morquio A assessed to be homozygous for a GALNS gene alteration for each of the growth phenotype categories and report that, as expected, gene alterations expected to severely affect protein function, such as deletions and nonsense gene alterations, are more common in patients with the most severely impacted growth phenotypes (Fig. 3, Supp. Table S4). We limited our analysis to patients reported to be homozygous for a GALNS gene alteration to avoid the possibility of any interallelic interactions (for examples from other disorders, see refs. Caciotti et al., 2003; Caciotti et al., 2005; Gieselmann et al., 1991; Thomas, 1994; Zlotogora and Bach, 1983]. The International Morquio A Registry documented the heterogeneity of Morquio A disease and reported that 68.4% of patients in the registry had a severe growth-impacted phenotype and that 25% had either a mild or attenuated growth phenotype [Montaño et al., 2007]. The analysis here, performed on an independent data set and reporting assessed growth phenotype, also found a similar proportion of growth-impacted severity phenotypes: of the patients with Morquio A homozygous for a GALNS gene alteration, 77% (107 of 139) have a reported severe growth phenotype, 4% (6 of 139) have an intermediate growth phenotype, and 19% (26 of 139) have a less severe growth phenotype. Missense alterations were the most commonly reported gene alteration type from homozygous patients across all growth phenotypes, in agreement with the high frequency of missense alterations reported in patients with Morquio A (Fig. 2B, Tomatsu et al. 2005b, Hendriksz et al., 2013a]. In addition to missense alterations, homozygous patients with the most severe growth phenotypes had other GALNS gene alteration types, including insertions and/or deletions, intronic alterations, and nonsense alterations. The missense alterations found in patients with more severe growth phenotypes are likely to be more deleterious alleles, with stronger possible effects on the GALNS enzyme (Supp. Table S4). With one exception, homozygous patients with less severe growth phenotypes had missense alterations; one patient with a less severe growth phenotype was reported to be homozygous for the intronic alteration c.898+1G>C [Tomatsu et al., 2005b]. Overall, these results are consistent with a previous independent analysis that found that most GALNS gene alterations associated with a less severe growth phenotype were missense alterations (Tomatsu et al., 2005b].
Locus-Specific Database
We have developed a Locus-Specific Database (LSDB) for the GALNS gene and a Website (http://galns.mutdb.org/) that supports browsing, searching, and sorting the database as well as submissions of new variations. Currently, the database includes previously published variants reported as “pathogenic” as well as those extracted from dbSNP [Sherry et al., 2001], SwissProt (UniProt Consortium, 2014), and Cosmic [Forbes et al., 2011]. BioMarin assisted in the collation of these mutations, but no mutations were from data collected as part of any BioMarin-sponsored clinical trial. This is in part because most BioMarin studies did not collect molecular testing data as part of uniform enrollment criteria and also because the scope of this update is to report only on published Morquio A mutations. We have converted all variants to the nomenclature recommended by HGVS [den Dunnen and Antonarakis, 2000], mapped transcript positions to the protein when the annotations were missing in the original publication as well as from the protein to the transcript when this annotation was missing (for variants present in SwissProt for example). We have also annotated variants as “reported pathogenic” or “benign” when a gene alteration was defined as such in a published article or as a “variant of unknown clinical significance” otherwise. We record additional curator comments about each gene alteration in a free text format in the comment section, which can include growth phenotype severity or other evidence of pathogenicity.
For missense alterations, we provide a bioinformatics analysis via MutPred [Li et al., 2009] and PolyPhen2 [Adzhubei et al., 2010] prediction scores and associated classifications for research purposes to aid assessment as to whether an amino acid substitution is potentially deleterious. These scores and classifications are for research use only and are not intended for use in clinical diagnosis of Morquio A. A MutPred score cutoff of 0.5 was used to classify variations as “predicted non-pathogenic” or “predicted pathogenic.” We have also listed every missense allele with either a MutPred or a PolyPhen2 score below 0.75 (Supp. Table S5). To aid in assessment of these alleles, the number of alleles reported and an estimate of the likelihood of a familial relationship between the reported patients is also provided.
In addition, we provide a JMOL protein structure viewer (Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/, accessed February 2014) for visualization of each gene alteration. All submissions will be carefully curated moving forward by our curation team. BioMarin has not and will not curate the LSDB. All GALNS variants in the LSDB are available for inclusion in other resources. This LSDB for GALNS gene alterations will encourage reporting of detected GALNS variants and will be a useful resource for the Morquio A community. Again, diagnosis of Morquio A should be made clinically and via demonstration of an enzyme defect in GALNS alongside other normal lysosomal enzyme activities in the context of a consistent clinical picture [Wood et al., 2013].
Future Prospects
Animal Models and Therapies
It is important to consider the use of GALNS molecular testing in the context of both what is known about Morquio A syndrome to date as well as in the context of Morquio A patient care. Much of our understanding about inherited disorders benefits from understanding animal models. However, although there are multiple mouse models carrying different mutations in the murine GALNS homolog [Tomatsu et al., 2003; Tomatsu et al., 2005a; Tomatsu et al., 2007; Tomatsu et al., 2008; Tomatsu et al., 2010], mouse and human catabolism of KS differs and the murine model did not develop the dysostosis phenotype of Morquio A [Tomatsu et al., 2010]. Despite the lack of a strong mouse disease model, enzyme replacement of recombinant human GALNS for Morquio A was able to proceed into clinical development, and recent clinical investigation has largely been focused on the evaluation of enzyme replacement therapy in Morquio A (Clinicaltrials.gov;[Hendriksz et al., 2013b]. Enzyme replacement therapy with elosulfase alfa (BioMarin Pharmaceutical Inc.) is now approved by the United States Food and Drug Administration for use in patients with Morquio A.
Although in some cases hematopoietic stem cell transplantation (HSCT) may play a role in other MPS disorders, there have been relatively few reports of HSCT performed in patients with Morquio A [Tomatsu et al., 2011; Valayannopoulos and Wijburg, 2011]. Additionally, chaperone therapies and other small molecule therapies, possibly designed with the aid of the GALNS structure [Rivera-Colón et al., 2012], could potentially benefit patients with Morquio A at some point in the future [Valayannopoulos and Wijburg, 2011], but useful application of these therapies would likely depend on accurate identification of the underlying molecular defect in patients with Morquio A, emphasizing the need for accurate molecular testing.
Newborn Screening
Once a therapy for a disorder becomes available, integration of newborn screening may be considered. GALNS molecular testing may be an important confirming test in the context of newborn screening. In the future it may be possible to routinely screen newborns for mucopolysaccharidoses disorders, including Morquio A, by using a liquid chromatography tandem mass spectrometry (LC–MS/MS) approach to detect reduced GALNS enzyme activity in dried blood spot samples [Spacil et al., 2013]. As the mucopolysaccharidoses are relatively rare disorders that can be difficult to diagnosis [Cimaz et al., 2009], widespread screening would be of great benefit. Additionally, the rapid proliferation and future clinical use of next-generation sequencing technologies may increase identification of individuals early in life with biallelic missense variants in GALNS prior to the development of a clear-cut clinical phenotype. These efforts and increasingly widespread availability of cheaper sequencing technologies in the future may lead to opportunities for earlier diagnosis of Morquio A patients; however, there are limitations to running molecular screening panels for the purpose of diagnosis that are outside of the scope of this review. We would like to re-emphasize that for Morquio A, and other similar monogenetic metabolic disorders for which there may be substantial numbers of “novel” or “private” gene alterations identified, enzyme activity testing is the standard for diagnosis [Wood et al., 2013].
Biomarkers
Although we recorded reported growth phenotype in the LSDB, we confirm, as previously reported in the last mutation update, that assessing genotype–phenotype relationships in Morquio A remains a challenge [Tomatsu et al., 2005b]. The heterogeneous alterations in the GALNS gene cause a spectrum of Morquio A disease manifestations and existing metrics of disease state do not always capture the full disease burden for patients [Harmatz et al., 2013; Montaño et al., 2007; Yasuda et al., 2013]. Biomarkers are needed to better understand Morquio A disease burden, aid patient counseling and the design of clinical evaluation plans. The levels of KS present in either plasma or urine have both been proposed as possible biomarkers for Morquio A disease severity [Dung et al., 2013; Harmatz et al., 2013]. The usefulness of plasma KS level as a biomarker is limited by the overlap between measurements from normal individuals and Morquio A patients [Longdon and Pennock, 1979; Martell et al., 2011; Zanetti et al., 2009]. Urinary KS levels from patients with Morquio A negatively correlate with both growth rates and final height and positively correlate with greater impairment in clinical tests of endurance/mobility and respiratory capacity [Northover et al., 1996; Montaño et al., 2008; Harmatz et al., 2013]. Future study into the relationship between possible biomarkers for Morquio A disease burden in the context of genotype–phenotype relationship could provide improved early diagnosis and/or improved clinical management of Morquio A syndrome.
Clinical and Diagnostic Strategies
Molecular analysis of the GALNS gene plays an important supporting role in the diagnosis of Morquio A. The gold standard for diagnosis of Morquio A is detection of deficient GALNS enzyme activity together with the wild-type activity of appropriate control enzymes [Wood et al., 2013]; detection of alterations in both GALNS alleles can confirm the diagnosis and aid genetic counseling. Accurate diagnosis of patients with Morquio A can be challenging, particularly for individuals with more slowly progressing disease, which makes confirmation of disease diagnosis by molecular analysis valuable.
The proportion of patients with Morquio A assessed as being homozygous for a GALNS gene alteration is substantial. Of the published genotypes from patients with Morquio A for whom at least one GALNS gene alteration was identified, 48% (257 out of 541) are assessed as being homozygous. Given the large number of alterations in the GALNS gene and that the three most frequently detected disease-associated alleles represent a total of 14% (152 out of 1,091) of all detected alleles, the 48% rate of assessed homozygosity suggests that factors such as founder effects and consanguinity may influence the high rate of assessed homozygous patient genotypes. It is possible that uniparental isodisomy (UPD) may cause homozygosity for a GALNS alteration in a small subset of cases. Although identification of these cases is important for genetic counseling, to date only one example has been reported [Catarzi et al., 2012].
Potentially, the percentage of patients with Morquio A assessed as being homozygous may be higher than the true frequency. In a subset of apparently homozygous patients, it is possible that deletions, rearrangements, and point mutations that eliminate binding sites for genotyping primers will cause allele dropout and a false assessment of apparent homozygosity. Of note, few large deletions have been reported from patients with Morquio A. Without additional deletion-duplication testing (by use of an appropriate technique such as quantitative genomic PCR (QF-PCR)), any deletion not entirely contained within an individual PCR amplicon will likely be missed by current sequencing approaches. Potentially, undetected large deletions could both cause apparent homozygosity and may contribute to the 13% of published patient genotypes for which only one GALNS gene alteration is identified (Fig. 2A).
Routine parental genotyping is recommended in the Morquio A diagnostic algorithm [Wood et al., 2013]; however, parental genotyping is not often performed and is even less frequently described in the literature. Parental genotyping results can aid interpretation of results from the patient and facilitate genetic counseling by allowing detection of situations in which the patient's results alone might lead to inaccurate conclusions being drawn; examples include cases in which two detected gene alterations are in cis, homozygosity due to UPD [Catarzi et al., 2012], and cases of allele dropout causing apparent homozygosity [Landsverk et al., 2012; Tylee et al., 2013]. One center performing molecular testing for multiple different autosomal recessive disorders found that, after performing parental testing on 75 patients initially assessed to be homozygous, four patients were incorrectly assessed as homozygotes due to allele dropout and two patients were genuinely homozygous but via UPD [Landsverk et al., 2012]. However, circumstances will not always allow for parental testing to be performed, particularly when it may raise a question of paternity.
The numerous GALNS gene alterations associated with Morquio A can make interpretation of molecular analysis results difficult. As with other Mendelian disease genes, the detection of novel GALNS alterations will occur relatively frequently. Additionally, there are GALNS sequence variants in exons (Supp. Table S6) and introns that have been reported without accompanying phenotypic information or a clear association with Morquio A. Although the functional consequences of some GALNS gene alterations can readily be predicted, such as nonsense alterations or large deletions, predicting the consequences of novel missense alterations from the sequence change alone can be difficult. The detection of a variant multiple times in patients with Morquio A from independent families strongly supports linking a variant with Morquio A disease; conversely, novel, difficult-to-interpret gene alterations (e.g., many missense alterations) should be interpreted with caution and only limited conclusions drawn [Morrone et al., 2014]. Consequently, reporting detected GALNS alterations facilitates the differentiation of disease-associated alleles from benign alterations and should be encouraged.
Acknowledgments
We would like to thank multiple researchers and laboratories for sharing data prior to publication and for discussions that helped motivate this manuscript: Karen Tylee and Heather Church (Willink Biochemical Genetics, Central Manchester University Hospitals NHS Foundation Trust, Saint Mary's Hospital Oxford Road, Manchester, United Kingdom); Moeen Al-Sayed (Department of Medical Genetics, King Faisal Specialist Hospital and Research Center, Riyadh, Saudi Arabia); Ana Carolina Brusius-Facchin, Francyne Kubaski, and Sandra Leistner-Segal (Laboratório de Genética Molecular, Serviço de Genética Médica, Hospital de Clínicas de Porto Alegre, Porto Alegre, Brazil); Michael Fietz (SA Pathology, Women's and Children's Hospital, North Adelaide SA, Australia), M. Jose Coll, Laura Gort, and Sonia Pajares (Sección de Errores Congénitos del Metabolismo-IBC, Servicio de Bioquímica y Genética Molecular, Hospital Clínic, CIBERER, IDIBAPS, Barcelona, Spain), Madhuri Hegde (Emory Genetics Laboratory, Emory University School of Medicine, Atlanta, Georgia, United States), Lucia Lacerda, Francisco Laranjeira, and Isaura Ribeiro (Unidade de Bioquímica Genética, Centro de Genética Médica Jacinto Magalhães do Centro Hospitalar do Porto, Porto, Portugal), Laura Pollard (Biochemical Genetics Laboratory, Greenwood Genetic Center, Greenwood, South Carolina, United States), and Raymond Wang (Children's Hospital of Orange County, Orange, California, United States). We thank Emanuela Izzo (BioMarin Pharmaceutical, Inc.) for review of the manuscript. We would also like to acknowledge the outstanding, seminal contributions of Shunji Tomatsu (University of Delaware, Wilmington, Delaware, United States) to both the broader mucopolysaccharidosis field and to the study of Morquio A syndrome.
Disclosure statement: Dr Mooney serves as a consultant for BioMarin Pharmaceutical. Dr Harmatz has received consultant fees and an honorarium from BioMarin Pharmaceutical and has been a study investigator. Dr Morrone has received consultant fees and limited travel support from BioMarin Pharmaceutical. Mrs Davidson, Dr Mealiffe, and Dr Miller are employees of BioMarin Pharmaceutical, and Mrs Ryles has worked for BioMarin Pharmaceutical. Dr Zawadzki is an employee of Health Interactions, which has contracts with BioMarin Pharmaceutical. Drs Atwood, Caciotti, Du, Francis-Lyon, and Oron declare no potential conflicts of interest.




