Structural Genomic Variation as Risk Factor for Idiopathic Recurrent Miscarriage
Contract grant sponsors: Wellcome Trust International Senior Research Fellowship (070191/Z/03/A) in Biomedical Science in Central Europe; Estonian Ministry of Education and Research Core Grants (SF0180022s12, SF0180026s09, SF0180142s08); European Union through the European Regional Development Fund (project HAPPY PREGNANCY, 3.2.0701.12-0047); Estonian Science Foundation Grant (ETF9030); EU ERDF through the Estonian Centre of Excellence in Genomics; European Commission FP7 project (gEUVADIS, 261123).
Communicated by Jacques Beckmann
ABSTRACT
Recurrent miscarriage (RM) is a multifactorial disorder with acknowledged genetic heritability that affects ∼3% of couples aiming at childbirth. As copy number variants (CNVs) have been shown to contribute to reproductive disease susceptibility, we aimed to describe genome-wide profile of CNVs and identify common rearrangements modulating risk to RM. Genome-wide screening of Estonian RM patients and fertile controls identified excessive cumulative burden of CNVs (5.4 and 6.1 Mb per genome) in two RM cases possibly increasing their individual disease risk. Functional profiling of all rearranged genes within RM study group revealed significant enrichment of loci related to innate immunity and immunoregulatory pathways essential for immune tolerance at fetomaternal interface. As a major finding, we report a multicopy duplication (61.6 kb) at 5p13.3 conferring increased maternal risk to RM in Estonia and Denmark (meta-analysis, n = 309/205, odds ratio = 4.82, P = 0.012). Comparison to Estonian population-based cohort (total, n = 1000) confirmed the risk for Estonian female cases (P = 7.9 × 10−4). Datasets of four cohorts from the Database of Genomic Variants (total, n = 5,846 subjects) exhibited similar low duplication prevalence worldwide (0.7%–1.2%) compared to RM cases of this study (6.6%–7.5%). The CNV disrupts PDZD2 and GOLPH3 genes predominantly expressed in placenta and it may represent a novel risk factor for pregnancy complications.
Introduction
Miscarriage is the most common pregnancy complication affecting up to 15% of all clinically recognized pregnancies [Stirrat, 1990]. The risk of miscarriage increases with age and is enhanced by a trend in developed countries to postpone childbearing to late 30s and early 40s with declined fertility rates [Nybo Andersen et al., 2000; Group TECW, 2010]. Recurrent miscarriage (RM), defined as ≥3 consecutive pregnancy losses before 22nd gestational week, is a heterogeneous disorder affecting up to 3% of couples aiming at childbirth [Christiansen et al., 2008]. It is a distressing condition for affected couples as each subsequent miscarriage leads to elevated risk of experiencing further pregnancy loss [Ogasawara et al., 2000] and increased probability of other pregnancy complications such as preterm delivery or small for gestational age newborns [Jivraj et al., 2001; van Oppenraaij et al., 2009]. Although a spectrum of factors is known to increase the risk to RM, including immune system dysfunction and thrombophilic disorders, the underlying etiology remains undetermined in about half of the cases [Rai and Regan, 2006; Allison and Schust, 2009].
A familial segregation of RM has been observed highlighting the contribution of the genetic predisposition to the disease [Christiansen et al., 1990; Kolte et al., 2011]. So far, majority of the studies focusing on the genetic susceptibility to RM have addressed single nucleotide polymorphisms (SNP) in more than 100 RM candidate genes, often providing conflicting results and no identified risk variants with confirmed strong effect [Nagirnaja et al., 2012; Rull et al., 2012, 2013]. More recently, paternal DNA fragmentation, epigenetic disturbances, and DNA copy number variants (CNVs) have been suggested to modify predisposition to RM [Rajcan-Separovic et al., 2010; Uuskula et al., 2010; Robinson et al., 2012]. CNVs represent deletions or duplications of >50 bp DNA sequence that disrupt around 13% of RefSeq genes [McCarroll et al., 2008; Zhang et al., 2009; Conrad et al., 2010]. It has been estimated that 17.7% of variability in gene expression may be attributed to these DNA variants and, subsequently, a growing number of genic CNVs and increased global burden of CNVs have been linked to several complex disorders [Stranger et al., 2007; Zhang et al., 2009; Girirajan et al., 2011]. Studies focusing on structural variation predisposing to pregnancy complications have been scarce and represent single reports addressing disease-associated CNVs in unexplained stillbirths and preeclampsia [Harris et al., 2011; Zhao et al., 2012, 2013]. A single study has addressed the contribution of CNVs in RM using array comparative genomic hybridization to screen 26 placentas from RM pregnancies for rare CNVs to identify candidate genes, which could be causative for the specific miscarriage [Rajcan-Separovic et al., 2010].
In the current genome-wide study, we set forward to address the role of CNVs in predisposing to RM among the couples of unexplained RM from Estonia and Denmark. The collaborative effort involved 558 RM cases with ≥3 consecutive miscarriages (in total, 309 RM female and 249 male partners), fertile multiparous control women (n = 205), and population-based samples from Estonian Biobank, Estonian Genome Center, University of Tartu (EGCUT; n = 1,000). We aimed to (A) assess the contribution of genome-wide burden of CNVs and (B) identify and characterize novel common CNVs in modulating the risk of RM (Fig. 1). We determined excessive cumulative burden of all CNVs (5.4 and 6.1 Mb per genome) in two RM cases possibly increasing their individual risk of the disease. The genomic CNV profile within the RM study group was significantly enriched for CNVs predominantly affecting genes related to innate immunity signaling and immunoregulatory interactions. As a major finding, we identified a multicopy duplication CNV (61.6 kb) at 5p13.3 conferring high, almost fivefold increased maternal risk of RM and disrupting two novel RM candidate genes, PDZD2 and GOLPH3, associated with pregnancy maintenance for the first time.

Materials and Methods
Study Subjects
RM cases and fertile control samples
RM cases and fertile controls have been recruited in two North European countries Estonia and Denmark, and the applied sample sets have been well characterized and previously exploited in RM research [Kruse et al., 2004; Rull et al., 2008, 2013; Kolte et al., 2011]. The study was approved by the Ethics Review Committee on Human Research of the University of Tartu, Estonia, and the Danish Central National Committee on Biomedical Research Ethics.
As both maternal and paternal genetic information determine the development of the fetus and the placenta, as well as the outcome of a pregnancy, the patient group consisted of female patients and their male partners experiencing idiopathic RM (≥3 consecutive miscarriages before week 22 of gestation without any identified cause). In total, the current study included 558 RM patients from Estonia (80 female and 39 male partners) and Denmark (229 female and 210 male cases) (Supp. Table S1). For all recruited cases, known clinical risk factors of RM have been excluded (Supp. Materials and Methods). The female patients were further subphenotyped as either primary (Estonia, n = 46; Denmark, n = 113) or secondary RM (Estonia, n = 34; Denmark, n = 116) based on the occurrence of consecutive miscarriages either before (if any) live births or following one or more live births, respectively.
The control group consisted of 205 fertile females from Estonia (n = 90) and Denmark (n = 115) with no history of miscarriages and at least two (Denmark) or three (Estonia) successful pregnancies ending with live birth (Supp. Table S1). Male partners of fertile female controls were not included into the study due to limited information on their previous reproductive history.
Population-based cohort samples from Estonian Biobank (EGCUT)
The carrier status of the identified RM-associated risk CNV at 5p13.3 was additionally determined for population-based cohort samples (n = 1,000; 504 men, 496 women) drawn from the Estonian population-based Biobank (EGCUT; www.biobank.ee; Supp. Materials and Methods) and previously subjected to SNP genotyping with Human370CNV-Duo SNP array (Illumina Inc., San Diego, CA) [Nelis et al., 2009] and genome-wide CNV calling (Priit Palta, unpublished data).
Discovery Phase: Genome-Wide SNP Genotyping and CNV Detection
The study design is schematically outlined in Figure 1. In the discovery phase, proportionally one-third subsample (n = 70; fertile controls, n = 27; RM cases, n = 43; Supp. Table S2) of all Estonian study subjects (n = 209) was genotyped using Illumina Human370CNV-Quad SNP array (Genotyping Core Facility, Estonian Biocentre). For each sample, calling of CNVs from the resulting genome-wide genotyping data was performed in parallel with two algorithms, QuantiSNP and PennCNV [Colella et al., 2007; Wang et al., 2007] (Supp. Materials and Methods). The initial CNV calls from the two algorithms were merged and only CNVs that were called by both algorithms for the same individual in the same genomic loci were considered in the subsequent global analysis. Discrete CNV regions (CNVRs) were defined by merging overlapping CNV calls across all individuals in a study group as described previously [Redon et al., 2006; Perry et al., 2008]. Genomic distribution of identified CNVRs was visualized using Circos software (http://circos.ca/) [Krzywinski et al., 2009]. The genome-wide CNV data of this study have been submitted to the Database of Genomic Variants (DGV; http://projects.tcag.ca/variation/).
For the EGCUT population-based cohort samples, the microarray data were processed in identical manner using parallel analysis of QuantiSNP and PennCNV and by considering only CNV calls that were made by both algorithms. Accurate CNV copy number for the 5p13.3 duplication locus was drawn from the QuantiSNP data due to its higher precision in copy number estimation in this locus.
Functional Enrichment Analysis
In order to acquire the up-to-date genomic annotation data for functional enrichment analysis, all CNVR breakpoint coordinates were converted to the latest version of the human reference sequence (from NCBI36/hg18 to GRCh37/hg19) (Supp. Materials and Methods). The list of genes within the identified CNV regions (extended by 10 kb on either side of the CNVR) was then acquired from the Ensembl database (version 69; http://www.ensembl.org/index.html). Functional enrichment analysis of subsequent gene sets was carried out separately for fertile controls and RM patients using g:Profiler gGOSt web-based software (http://biit.cs.ut.ee/gprofiler/) [Reimand et al., 2007, 2011] (Supp. Materials and Methods). Two outlier cases with increased genomic burden of CNVs (RM-M45 and RM-F4) were excluded from the analysis to avoid biased results. Results of Gene Ontology (GO) and Reactome (REAC) datasets with up to third relative hierarchy level were taken into account and enrichment for functional terms was considered significant if the multiple testing corrected enrichment P-value was <0.05.
Prioritization of CNVRs for Experimental Confirmation and Subsequent Analysis
Discrete CNV regions identified in the discovery phase based on the whole genome SNP array genotyping data (full list of CNVRs in Supp. Table S3) were selected for experimental confirmation and further analysis if the following criteria were met: CNVR was (1) present in >1 individual (criterion met by 118 out of 423 nonoverlapping CNVRs), (2) found only among RM patients or overrepresented in RM patients with odds ratio (OR) ≥1.5 (45/118 CNVRs), and (3) intersected with or located in the proximity (up to approximately 200 kb) of biological candidate genes with a potential impact on the course of pregnancy based on previously published literature (9/45 CNVRs). As our study aimed to identify common rearrangements predisposing to RM, previous reports in the DGV database for the prioritized CNVRs of this study were considered as confirmation of true CNV loci, rather than as an exclusion criterion due to unknown reproductive success of the genotyped individuals in these reports.
Experimental Copy Number Estimation of Prioritized CNVRs Using TaqMan qPCR
For experimental testing, TaqMan quantitative PCR (qPCR) was performed with one assay for seven of the prioritized CNVRs and with two assays in parallel for the two largest CNV loci as described in Supp. Materials and Methods (Supp. Table S4). Ten nanograms of genomic DNA was amplified using predesigned TaqMan copy number assays (Applied Biosystems, Foster City, CA; Supp. Table S5) or previously published TaqMan qPCR primers and FAM-tagged probe (6p21.33 CNVR; Supp. Table S6) [Parajes et al., 2007]. Copy number was normalized to the reference RNase P and population-specific pool of control DNAs (Supp. Materials and Methods). The diploid genomic copy number was calculated by multiplying the normalized TaqMan qPCR copy number estimates by two. Because of limitations of TaqMan qPCR assay to accurately determine very high diploid copy numbers, individuals with estimated locus copy number larger than four were assigned into copy number class “>4 copies per diploid genome.”
Experimental Fine Mapping and Characterization of the 5p13.3 Rearrangement
The confirmation of the 5p13.3 duplication endpoints estimated by SNP array was performed with four EvaGreen qPCR assays (Supp. Table S6; Supp. Fig. S1) flanking the predicted breakpoints and using samples with known 5p13.3 copy number based on the data of TaqMan qPCR copy number typing (Supp. Materials and Methods). Copy number was estimated using absolute quantification method and normalized to the reference gene ALB and population-specific pool of control DNAs.
The exact position of duplication breakpoint junction in three 5p13.3 duplication carriers was determined using DNA sequencing by primer walking and targeting breakpoint junction region (5.8 kb) defined based on EvaGreen mapping (Supp. Materials and Methods). A control PCR spanning the identified breakpoint junction of the 5p13.3 tandem duplication (BP-PCR) was applied to confirm the duplication carriership in all Estonian and Danish RM case-control samples previously addressed with TaqMan qPCR (Supp. Materials and Methods).
The DNA sequences flanking the identified duplication breakpoints (±1,000 bp) were screened for repetitive elements using RepeatMasker software (http://www.repeatmasker.org/) and searched for the non-B DNA motifs including direct and inverted repeats, short-tandem repeats, and cruciform motifs using non-B DNA motifs search tool (http://nonb.abcc.ncifcrf.gov/apps/nBMST/default/).
Expression Profile Analysis of PDZD2 and GOLPH3 Genes
The expression analysis was performed using human tissue cDNA panels Human MTC panel I and II (BD Biosciences Clontech, Palo Alto, CA) consisting of pools of samples for each tissue (Supp. Materials and Methods). The expression profile of the GOLPH3 and PDZD2 genes was determined with TaqMan qPCR approach using predesigned TaqMan gene expression assays (Applied Biosystems) and normalization to the reference transcript of HPRT.
Copy Number Assignment of TaqMan qPCR Values
The copy number assignment for simple deletion and/or duplication polymorphisms was performed manually, whereas for the multicopy 5p13.3 locus, average copy number ratio of two TaqMan assays located within the rearranged region was used for the assignment of each sample into a distinct copy number cluster with k-means clustering method in the statistical package R (ver. 2.15.0; http://www.R-project.org/) (Supp. Materials and Methods). The resulting grouping was confirmed using EvaGreen qPCR performed with assay “EvaGr assay 2” located within the rearranged region (Supp. Fig. S1). Furthermore, the 5p13.3 duplication carriership was confirmed in all Estonian and Danish RM cases and fertile controls using the duplication junction-specific control PCR (BP-PCR). Subsequent groups of carriers versus noncarriers were subject to genetic association testing (Supp. Fig. S2).
Genetic Association Testing
Genetic association of the 5p13.3 duplication with the clinical diagnosis of RM was tested using logistic regression model in the statistical package R (version 2.15.0) in the Estonian and Danish RM cases/fertile control sample sets with. Subjects were assigned as either carriers or noncarriers (wild type) of the tandem duplication. The obtained population-specific results were subsequently combined in the classical meta-analysis approach based on effect size estimates (beta-statistic) and standard errors and performed with the inverse-variance method under fixed-effects model in the statistical package R. Alternatively, to correct for the asymmetric case/control ratio in the Estonian and Danish sample sets, a z-score-based meta-analysis was performed combining the logistic regression P-values, effect directions, and effective sample sizes using METAL software as described by Willer et al. (2010). In Estonians, the RM patient group was further tested against Estonian Biobank population-based cohort samples using logistic regression analysis as described above. Results with P-values <0.05 were considered significant.
Results
Increased Genomic Burden of CNVs in Two RM Patients
In the discovery phase, the whole-genome screening (Illumina Human370CNV-Quad SNP array) for structural variants was performed in one-third subset of Estonian case-control samples (n = 70; Supp. Table S2). On average, 13.3 CNVs were determined per individual in RM cases (n = 43) and 12.6 CNVs in fertile female controls (n = 27). Among all detected CNVs (n = 915) in the full discovery sample, a 2.1-fold excess of deletions compared to duplications was observed consistent with previously reported ratios [Redon et al., 2006; Conrad et al., 2010]. However, the median length of duplications exceeded significantly the size of deletions (60.8 kb vs. 26.1 kb; Mann–Whitney U test, P = 1.82 × 10−13).
Genomic burden analysis of all detected CNVs per individual revealed two outlier RM patients RM-F4 and RM-M45 with increased cumulative size of CNVs exceeding the rest of the RM cases more than fivefold (6.1 Mb and 5.4 Mb vs. median of 1.0 Mb, respectively) (Fig. 2; Supp. Fig. S3). The case RM-F4 was the carrier of only heterozygous deletions (n = 19; Fig. 2, Supp. Fig. S4A) with the majority being longer than 100 kb (n = 15; median 313 kb) and deleting altogether 50 genes. Remarkably, the 5.4 Mb of CNVs (in total, n = 29; Fig. 2, Supp. Fig. S4A) in the case RM-M45 rearranged a total of 378 genes of which 113 duplicated and five deleted genes were located within the genomic region of immunoglobulin heavy chain (IGH) gene cluster at 14q32.33 potentially regulating the immune function during pregnancy. Additionally, heterozygous deletions of genes related to spermatogenesis (SOHLH1; MIM# 610224, SPATC1; MIM# 610874) [Goto et al., 2010; Suzuki et al., 2012] and known RM candidate genes (such as C4A (MIM# 120810), C4B (MIM# 120820), and IGF2 (MIM# 147470)) [Laitinen et al., 1991; Ostojic et al., 2008] were identified in this male patient potentially increasing the individual's risk of RM disease. Both patients were from couples suffering from unexplained primary RM with no prior live births at the time of recruitment and had experienced three (RM-M45) or four (RM-F4) consecutive miscarriages occurring before gestational week 12. The available medical records for the male partner RM-M45 do not report any other diseases. During the postrecruitment period, the female patient RM-F4 carrying 50 hemizygous genes has experienced further miscarriages (n = 3) but also live births (n = 2) as a result of clinical management. Additionally, she has developed insulin-dependent diabetes mellitus and euthyroid goiters potentially also attributable to the large cumulative burden of deletions.
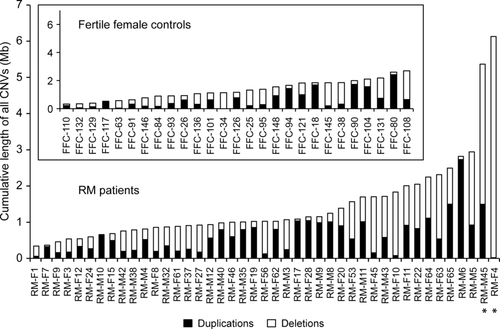
Functional Profiling of Genes Disrupted by CNVRs Reveals Enrichment of Immune Signaling Pathways among RM Cases
The CNVs identified in the discovery phase clustered into 423 nonoverlapping CNVRs in RM cases and fertile controls that were uniformly distributed across the genome (Supp. Table S3, Supp. Fig. S4B). Functional profiling analysis of all genes disrupted by these CNVRs in either RM patient group (excluding two outlier cases with increased CNV burden) or among controls (1151 and 553 genes, respectively) was undertaken to identify pathways and processes significantly modulated by the rearrangements. In RM patients, the analysis specifically highlighted the role of impaired immune signaling, antigen binding, and immunoregulatory interactions at the fetomaternal interface in recurrent pregnancy loss (Table 1). Notably, alterations in the REAC pathways of “innate immunity signaling” (9.3% of genes rearranged in the pathway, P = 9.15 × 10−3), “Complement cascade” (13.0% of genes, P = 1.11 × 10−3), and “Fc gamma receptors interact with antigen-bound IgG” (21.1% of genes rearranged by CNVs, P = 2.60 × 10−4) were specific to RM cases as none of the genes belonging to these categories were affected by structural variants in fertile controls (Table 1).
| Functional category | RM casesa | Controls | |||
|---|---|---|---|---|---|
| Type | ID | Name | Level | Corrected P-valueb | Corrected P-valueb |
| Functional categories significantly enriched in RM cases | |||||
| GO | 0003823 | Antigen binding | 1 | 1.16 × 10−4 | 1.00 |
| REAC | 168256 | Signaling in immune system | 1 | 1.44 × 10−3 | 1.00 |
| 168249 | Innate immunity signaling | 1.1 | 9.15 × 10−3 | No genesc | |
| 166658 | Complement cascade | 1.1.1 | 1.11 × 10−3 | No genesc | |
| 198933 | Immunoregulatory interactions between a lymphoid and a nonlymphoid cell | 1.2 | 1.61 × 10−4 | 1.00 | |
| 199161 | Fc gamma receptors interact with antigen-bound IgG | 1.2.1 | 2.60 × 10−4 | No genesc | |
| Functional categories significantly enriched in fertile controls | |||||
| GO | 0060089 | Molecular transducer activity | 1 | 1.00 | 2.19 × 10−11 |
| 0004871 | Signal transducer activity | 1.1 | 1.00 | 2.19 × 10−11 | |
| 0071944 | Cell periphery | 2 | 1.00 | 1.28 × 10−7 | |
| 0005886 | Plasma membrane | 3 | 1.00 | 4.17 × 10−8 | |
| 0044425 | Membrane part | 4 | 1.00 | 3.27 × 10−6 | |
| 0031224 | Intrinsic to membrane | 4.1 | 1.00 | 5.36 × 10−6 | |
| 0016021 | Integral to membrane | 4.1.1 | 1.00 | 5.36 × 10−5 | |
| 0016020 | Membrane | 5 | 1.00 | 3.90 × 10−4 | |
| 0004872 | Receptor activity | 6 | 1.00 | 1.16 × 10−15 | |
| 0038023 | Signaling receptor activity | 6.1 | 1.00 | 2.02 × 10−13 | |
| 0004888 | Transmembrane signaling receptor activity | 6.1.1 | 1.00 | 1.36 × 10−14 | |
| REAC | 372790 | Signaling by GPCR | 1 | 1.00 | 5.60 × 10−3 |
| 381753 | Olfactory signaling pathway | 1.1 | 1.00 | 9.99 × 10−7 | |
| 381750 | Olfactory receptor-G protein olfactory trimer complex formation | 1.1.1 | 1.00 | 9.99 × 10−7 | |
- The list of rearranged genes in either patients (n = 43) or controls (n = 27) was subjected to functional enrichment analysis using g:Profiler software (Reimand et al., 2007) and included Gene Ontology (GO) and Reactome (REAC) functional categories up to third relative hierarchical level. gProfiler performs statistical enrichment analysis to identify functional groups and/or biological pathways that are significantly overrepresented in the user-provided gene list.
- a Two outlier cases RM-F4 and RM-M45 with increased genomic burden of CNVs (Fig. 2) removed from the analysis.
- b Multiple testing corrected enrichment P-value.
- c None of the genes in this functional category were disrupted by CNVs among controls.
- GPCR, G protein coupled receptor.
Among the controls, the analyzed CNVRs likely represent benign rearrangements as only biological pathways of general cellular function were identified, including signaling of olfactory receptors (“olfactory signaling pathway,” REAC:381753, P = 9.99 × 10−7), a large class of genetically diverse proteins in humans known to be affected by CNVs in healthy individuals [Wong et al., 2007; Hasin-Brumshtein et al., 2009; Mills et al., 2011].
Experimental Testing of Prioritized CNVRs Detected by SNP Array
In order to identify common distinct CNV regions increasing the risk of RM, nine prioritized CNVRs were selected based on the genome-wide CNV screening results (Supp. Table S3; Supp. Materials and Methods) and subjected to copy number typing using TaqMan qPCR in the discovery sample set (n = 70) (see also Fig. 1). Precise copy number estimation by TaqMan qPCR assays was observed for four prioritized CNVRs, identifying all CNV carriers predicted based on microarray analysis with no false positives (Supp. Table S4, Supp. Fig. S5; false negative rate across regions 1.4%). Three CNV regions— IGKV (immunoglobulin kappa variable cluster at 2p11.2; MIM# 146980), DKK2 (Dickkopf 2 homolog at 4q25; MIM# 605415), and PDZD2:GOLPH3 (PDZ domain containing 2; MIM# 610697; golgi phosphoprotein 3 at 5p13.3; MIM# 612207)—were carried onto the next experimental stage to analyze the full Estonian RM case-control sample set using the established TaqMan qPCR approach (Fig. 1). SEPT14 CNVR (septin 14 at 7p11.2; MIM# 612140) was excluded from the next stage due to its restricted testis-specific expression [Peterson et al., 2007].
Although confirmed as copy number variable by TaqMan qPCR, precise locus copy number estimation of the remaining five prioritized CNVRs (Supp. Table S4) was most likely hindered due to complex genomic architecture and rearrangements of the region as also evident based on previous CNV studies (Supp. Fig. S6). Thus, these CNVRs were excluded from further genetic association testing.
Significantly Increased Prevalence of PDZD2:GOLPH3 Duplication in Estonian and Danish RM Cases
Among the three CNVRs (PDZD2:GOLPH3 duplication, IGKV deletion/duplication, and DKK2 deletion) tested in the full Estonian sample set (Supp. Table S1), PDZD2:GOLPH3 duplication detected in up to four copies per diploid genome exhibited the strongest effect (OR = 7.28) with a higher prevalence of duplication carriers among the RM cases compared to fertile controls (9/119, 7.6% vs. 1/90, 1.1%, respectively) (Supp. Table S7). The Replication study of the PDZD2:GOLPH3 CNVR performed in Danish RM cases and fertile controls (in total, n = 554; Supp. Table S1) confirmed the increased carrier frequency of the rearrangement (RM patients, 19/439, 4.3%; controls, 2/115, 1.7%; Table 2). However, a sex-stratified analysis of the Danish RM patients revealed the higher prevalence only among the female patients (6.6%) that was comparable to Estonian female cases (7.5%) (Fig. 3A, Table 2). The PDZD2:GOLPH3 duplication reached statistically significant association with an increased risk of RM (OR = 4.82, P = 0.012) in the meta-analysis combining the results of the Estonian and Danish female patient-control samples (in total, cases n = 309, controls n = 205). The high prevalence of the duplication observed among Estonian male RM partners was not replicated in Denmark (Supp. Table S8). The observed differences could be attributed to the different size of the Estonian and Danish sample sets (n = 208 and n = 558, respectively). These North European subjects have been recruited according to similar clinical criteria minimizing the interstudy heterogeneity and used in parallel in previous collaborative research [Nagirnaja et al., 2012; Rull et al., 2013].
| Women in association testing | Association testinga | |||||
|---|---|---|---|---|---|---|
| Controls | Number of subjects/carriers (%) | Female RM cases | Number of subjects/ carriers (%) | OR | CI (95%) | P-value |
| Association testing 1: full Estonian RM case-control sample | ||||||
| Fertile women | 90/1 (1.1) | All cases | 80/6 (7.5) | 7.22 | 0.85–61.25 | 0.070 |
| Primary RM | 46/4 (8.7) | 8.48 | 0.92–78.18 | 0.059 | ||
| Secondary RM | 34/2 (5.9) | 5.56 | 0.49–63.45 | 0.167 | ||
| Association testing 2: Estonian RM cases versus EGCUT cohort | ||||||
| EGCUT women | 496/5 (1.0) | All cases | 80/6 (7.5) | 7.96 | 2.37–26.75 | 7.9 × 10−4 |
| Primary RM | 46/4 (8.7) | 9.35 | 2.42–36.15 | 0.001 | ||
| Secondary RM | 34/2 (5.9) | 6.14 | 1.15–32.88 | 0.034 | ||
| Replication study: Danish RM case-control sample | ||||||
| Fertile women | 115/2 (1.7) | All cases | 229/15 (6.6) | 3.96 | 0.89–17.62 | 0.071 |
| Primary RM | 113/9 (8.0) | 4.89 | 1.03–23.15 | 0.045 | ||
| Secondary RM | 116/6 (5.2) | 3.08 | 0.61–15.60 | 0.174 | ||
| Meta-analysis: Estonian and Danish RM cases and fertile controls | ||||||
| Fertile women | 205/3 (1.5) | All cases | 309/21 (6.8) | 4.82 | 1.42–16.40 | 0.012 |
| Primary RM | 159/13 (8.2) | 5.86 | 1.64–20.94 | 0.007 | ||
| Secondary RM | 150/8 (5.3) | 3.70 | 0.96–14.25 | 0.058 | ||
- Association testing was performed with duplication carriers versus noncarriers as established based on TaqMan qPCR and confirmed by junction-spanning PCR for RM cases and fertile controls and estimated by QuantiSNP based on SNP genotyping and CNV calling data for EGCUT samples (Supp. Materials and Methods).
- a Association testing was performed using logistic regression analysis in Estonia and Denmark separately. The results were subsequently combined in the meta-analysis using inverse-variance method under fixed-effects model. Results with P-value <0.05 were considered significant and are indicated in bold.
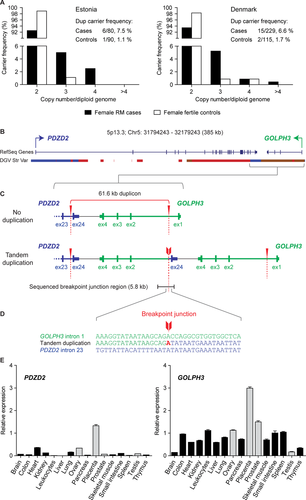
Notably, the strongest effect of the PDZD2:GOLPH3 CNV was detected among the subgroup of women with primary RM (Estonian cases: n = 46, CNV carrier frequency 8.7%; Danish, n = 113, 8.0%) defined as ≥3 consecutive miscarriages and no preceding live births (meta-analysis, n = 159; OR = 5.86, P = 0.007) (Table 2). A z-score-based meta-analysis correcting for the asymmetric case/control ratio provided consistent results in association testing including all female cases (P = 0.011) or only cases of primary RM (P = 0.006; Supp. Table S9).
The two CNVRs (IGKV locus, Del/Dup; DKK2 locus, Del) with small differences in carrier frequencies in the full Estonian case-control sample (Supp. Table S7) likely represent benign common copy number variation with no major effect on the RM phenotype in our study.
Low Prevalence of the PDZD2:GOLPH3 Duplication in Worldwide Cohorts
The high risk of RM associated with the PDZD2:GOLPH3 duplication was further confirmed when Estonian RM cases were compared to the Estonian Biobank (EGCUT) cohort samples (in total, n = 1,000) (Table 2). The most significant association among the subgroups of cases was observed for female patients tested against female cohort subjects (n = 496; prevalence 1.0%; OR = 7.96, P = 7.9 × 10−4). Importantly, the carrier frequency of the PDZD2:GOLPH3 duplication in the whole EGCUT cohort (9/1,000; 0.9%) was as low as observed among the Estonian and Danish controls (Table 3) and similar carrier frequency has been consistently reported for various large worldwide population-based cohorts in the DGV (sample size range: n = 776–2,026; in total, n = 5846; duplication carrier frequency from 0.7% to 1.2%) (Table 3).
| CNV | Number of | ||||||
|---|---|---|---|---|---|---|---|
| carriers | carriers/study | CNV | CNV size | ||||
| Populationa | (%) | group | Phenotype | Detection methodb | type | (kb) | Reference |
| Dataset of this study | |||||||
| Denmark | 1.7 | 2/115 | Fertile controls | TaqMan qPCR | Dup | 61.6 | This study |
| Estonia | 1.1 | 1/90 | Fertile controls | Illumina Human370CNV-Quad, TaqMan qPCR | Dup | 61.6 | This study |
| Estonia | 0.9 | 9/1,000 | Cohort | Illumina Human370CNV-Duo | Dup | 61.6 | This study; Nelis et al. (2009) |
| Dataset presented in the Database of Genomic Variants (DGV) | |||||||
| Ontario, Canadac | 0.7 | 8/1,190 | Cohort | Affymetrix 500K and 100K | Dup | 35.1; 62d | Zogopoulos et al. (2007) |
| Worldwide (HGDP, NINDS)e | 1.1 | 21/1,854 | Cohort; neurological disease controls | Illumina HumanHap300, Illumina HumanHap240S, Illumina HumanHap650Y, Illumina HumanHap550 | Dup | 68.1 | Itsara et al. (2009) |
| Worldwide (HapMap, PopGen)c, f | 1.2 | 9/776 | Healthy controls cohort | Affymetrix 500K | Dup | 35.1; 95; 146d | Pinto et al. (2007) |
| Philadelphia, PAc,g | 1.0 | 20/2,026 | Healthy controls cohort | Illumina HumanHap550 V1 | Dup | 28.5; 38.9; 71.7d | Shaikh et al. (2009) |
- a In case of multiple studies targeting an identical study group (e.g., HapMap collection), results for the largest sample collection are presented.
- b Only carrier frequencies from studies applying SNP array copy number estimations are included from the DGV database.
- c Data for multiple overlapping nonrecurrent CNVs in 5p13.3 identified in the study are merged.
- d CNV sizes of all overlapping CNVs in the region identified in the study.
- e HGDP: cohort of the Human Genome Diversity Panel, 51 world populations, n = 1,064; NINDS, neurological disease controls of European descent from National Institute for Neurological Disorders and Stroke, n = 790.
- f HapMap: healthy individuals from four populations—parent–offspring trios of the Yoruba from Nigeria (YRI; n = 30), parent–offspring trios of European descent from Utah (CEU; n = 30), unrelated Japanese from Tokyo, Japan (JPT; n = 45), and unrelated Han Chinese from Beijing, China (CHB; n = 45); PopGen: unrelated healthy individuals from Northern Schleswig-Holstein (Northern Germany), n = 506.
- g The cohort included 1,492 unrelated individuals, 80 mother–father–child trios, 325 mother–child and 140 father–child duos, 59 siblings, and 10 twins.
- Dup, duplication.
Fine-Scale Experimental Mapping of the PDZD2:GOLPH3 Duplication CNV
Experimental fine-scale mapping of the 5p13.3 duplication confirmed the extent of the CNV predicted by the discovery SNP microarray data from the last intron of the PDZD2 to the first intron of the GOLPH3 genes, which are transcribed from the opposite DNA strands (Fig. 3B). Subsequent sequencing of the breakpoint junction-spanning region in the PDZD2:GOLPH3 duplication carriers refined the exact genomic coordinates of the CNV as Chr5: 32106204 – 32167777 and length as 61.6 kb (compared to 52.4 kb based on SNP array estimation) (Fig. 3C). The presence of identical recurrent tandem duplication was detected in all North European carriers of the PDZD2:GOLPH3 CNV independent of the duplicon copy number.
The flanking genomic context of the PDZD2:GOLPH3 duplication endpoints were characterized by a high number of repetitive elements in the proximity and overlapping with both breakpoint sites (Supp. Fig. S7). The proximal duplication endpoint was located within the DNA repetitive sequence MER1A and the distal endpoint in the AluJb element, whereas no extensive DNA sequence homology between the two repeat elements was observed. Also, no DNA sequences leading to non-B DNA conformation were identified and only a microhomology of one nucleotide was determined at the junction of the duplication endpoints by sequencing (Fig. 3D). Thus, repeat-mediated rearrangement mechanisms other than nonallelic homologous recombination have possibly contributed to the occurrence of this recurrent duplication (reviewed in [Hastings et al., 2009]).
Expression Analysis of PDZD2 and GOLPH3 Genes in Human Tissues Reveals the Highest Expression in Placenta
Although GOLPH3 was ubiquitously expressed in all tested tissues (human MTC panels I and II), it exhibited the highest transcript level in the placenta (average expression relative to reference HPRT, 2.99 ± 0.06 SEM), followed by prostate, ovary, and leucocytes (relative expression 1.50 ± 0.05, 1.13 ± 0.02, and 1.12 ± 0.04; respectively) (Fig. 3E). In general, the level of PDZD2 transcripts was low in all tissues; however, the highest expression was also determined for the placenta exceeding fourfold the detected mRNA quantities in the two other sites of highest expression, heart and ovary (relative expression 1.33 ± 0.05 vs. 0.36 ± 0.01 and 0.34 ± 0.0, respectively) (Fig. 3E). Overall, the expression profile of these genes points to their functional relevance in the placenta and potentially in other reproductive organs.
Discussion
In this study, we have addressed the effect of genome-wide CNVs and contribution of selected CNV regions in modulating the predisposition to RM. The genome-wide CNV profiling revealed a significant enrichment of rearranged genes linked to innate immune signaling and immunoregulatory interactions within the RM study group, indicative of a potential effect on early pregnancy maintenance. As a main result, we report a novel duplication locus at 5p13.3 conferring high risk of RM among the female subjects of both Estonia and Denmark, and disrupting the PDZD2 and GOLPH3 genes predominantly expressed in placenta and associated with pregnancy maintenance for the first time.
The large cumulative burden of CNVs and specifically long deletions may modify an individual's predisposition to RM due to increased chances of disrupting specific key genes or pathways essential for early pregnancy maintenance. A case-by-case analysis performed in this study identified two RM cases RM-F4 and RM-M45 distinguished by heavy burden of CNVs and accumulation of several long deletions in RM-F4 (n = 15 out of 19 CNVs; Fig. 2). Several potential or known RM candidate genes that may independently or synergistically elevate the risk of RM were rearranged within these cases, including immunomodulatory loci such as 119 genes from the IGH cluster and the previously reported RM-associated C4A and C4B genes [Laitinen et al., 1991]. A subset of patients may thus exhibit increased risk of RM disease attributable to excessive genomic burden of CNVs; however, the finding remains to be confirmed by future studies.
The detailed functional enrichment analysis of genes under CNVs within the RM study group confirmed the highly specific overrepresentation of pathways related to immune function and highlighted the processes most sensitive to CNV alterations in early pregnancy such as antigen binding, immunoregulatory interactions, innate immunity signaling, and complement cascade pathway associated with RM previously (Table 1) [Li and Huang, 2009; Denny et al., 2013]. The CNV profile modifying the repertoire of presented antigens and alloimmune responses may affect the subtle balance of the immunological tolerance at the fetomaternal interface. As mother is carrying a semiallogeneic fetus expressing paternally inherited alloantigens, maternal immune rejection has been implicated in the etiology of preeclampsia and RM previously [Wilczynski, 2006; Guleria and Sayegh, 2007; Faridi and Agrawal, 2011]. The major contribution of fetomaternal immune function in the development of RM has also been demonstrated in gene expression profiling of the maternal decidual tissue and chorionic villi of aborted fetuses [Baek et al., 2002; Krieg et al., 2012].
As a major outcome of this study, a common multicopy duplication at 5p13.3 was identified conferring increased maternal risk to RM in North European populations, Estonians and Danes (meta-analysis OR = 4.82, P = 0.012). The prevalence of the PDZD2:GOLPH3 CNV was detected with more than fivefold higher frequency in the female RM patients compared to fertile women (6.6%–7.5% vs. 1.1%–1.7%) or worldwide population cohorts (0.7%–1.2%; Tables 2 and 3). The largest effect of GOLPH3:PDZD2 duplication as a risk factor was detected among the subgroup of women diagnosed with primary RM (meta-analysis, OR = 5.86, P = 0.007; Table 2). The duplication breakpoints of the identified PDZD2:GOLPH3 recurrent CNV (61.6 kb in size) occurring in up to >4 diploid copies (Fig. 3A) were positioned within the PDZD2 and GOLPH3 genes transcribed in the opposite directions (Fig. 3C). Although the 5p13.3 duplication does not directly alter the copy number of entire coding regions of the genes, the modifications in the local genomic context may nevertheless interfere with the transcription leading to dysregulation of the involved or neighboring genes as reported previously [Henrichsen et al., 2009a, 2009b]. The potential functional relevance of the identified rearrangement in pregnancy was supported by the expression profiling of PDZD2 and GOLPH3 in the human tissue panels revealing mutually high transcription levels in the placenta (two to fourfold increase compared to other tissues) but also in the maternal reproductive organ ovary (Fig. 3E). We speculate that the significantly increased predisposition to RM among the female duplication carriers may be attributed to the joint effect in maternal reproductive tissues and in the placental tissue carrying a maternally inherited duplication CNV. The mechanism of action and joint effect of the 5p13.3 CNV remains to be addressed further due to current limited availability of ovarian and early miscarriage placental samples carrying the 5p13.3 CNV.
Most importantly, a recent study addressing the contribution of maternal CNVs in cases of preeclampsia (PE)—a severe late pregnancy disorder originating from placental dysfunction—identified the PDZD2:GOLPH3 duplication as the genetic risk factor of the disease [Zhao et al., 2013]. The reported 50.4 kb duplication colocalized with the CNV identified in the current study and occurred with the prevalence of 8.3% among the affected and 1.6%–1.9% among unaffected female subjects. The identified risk to PE (OR = 4.80, P < 0.05) is close to the effect detected for the RM susceptibility in this study (Table 2). The possible common origin of various pregnancy disorders has been acknowledged previously, including overlapping causality of RM and PE [Li and Huang, 2009; Baig et al., 2013]. Mutations in the dosage-sensitive gene GLUT3 (MIM# 138170) have been reported as the common genetic risk factor leading to either early pregnancy loss or fetal growth restriction [Ganguly et al., 2007]. Thus, it is plausible that the identified PDZD2:GOLPH3 duplication may represent a pleiotropic risk factor in the genetic etiology of not only RM but also other pregnancy complications.
Neither of the novel RM-associated genes PDZD2 or GOLPH3 has been linked to placental function or pregnancy maintenance previously. The function of PDZD2 is poorly defined and has mainly been addressed as a tumor suppressor [Yeung et al., 2003; Tam et al., 2006]. More extensively studied conserved GOLPH3 is essential for Golgi trafficking and maintenance of its structure [Dippold et al., 2009; Wood et al., 2012]. Recently, genomic amplification of the GOLPH3 gene was linked to oncogenic features [Scott et al., 2009; Wang et al., 2012; Hu et al., 2013] and was shown to activate the signaling pathway of mechanistic target of rapamycin (mTOR), an essential component of mammalian reproductive function [Murakami et al., 2004; Wen et al., 2005; Busch et al., 2009; Yu et al., 2011; Gonzalez et al., 2012; Tanwar et al., 2012]. Alterations in mTOR signaling have been associated with multiple reproductive disorders in mice and human, including RM [Roos et al., 2007; Adhikari et al., 2010; Hirota et al., 2011; Leconte et al., 2011; Vatin et al., 2012], thus providing the potential functional link between the amplification of GOLPH3 and development of RM disease.
In summary, the findings of this study highlight the genetic heterogeneity of RM not only by characterizing the potential predisposing effect of the cumulative burden of genome-wide CNVs, but also by identifying a common PDZD2:GOLPH3 duplication, as RM risk factor detected in two independent North European sample sets. Importantly, the identified PDZD2:GOLPH3 duplication may represent a novel genetic risk factor for several pregnancy complications. The underlying functional effect of the PDZD2:GOLPH3 duplication in reproductive organs, placenta, and potentially in tumorigenesis remains to be addressed.
Acknowledgments
We appreciate all the study participants from Estonia and Denmark. We are thankful to Professor Andres Metspalu for suggestions on the manuscript and providing genotyping data of population-based controls from the Estonian Biobank in the Estonian Genome Center of the University of Tartu. We acknowledge Dr. Jüri Reimand for advice on functional enrichment analysis.
Disclosure statement: The authors declare no conflict of interest.




