Assessment of Adenosine Triphosphatase Phospholipid Transporting 8B1 (ATP8B1) Function in Patients With Cholestasis With ATP8B1 Deficiency by Using Peripheral Blood Monocyte-Derived Macrophages
Supported by the Japan Agency for Medical Research and Development (grant numbers JP17ek0109116, JP19ek0109408, and JP19ek0109419 to H.H.).
The funding source did not participate in the study design or execution.
Potential conflict of interest: Nothing to report.
Abstract
Adenosine triphosphatase phospholipid transporting 8B1 (ATP8B1) deficiency, an ultrarare autosomal recessive liver disease, includes severe and mild clinical forms, referred to as progressive familial intrahepatic cholestasis type 1 (PFIC1) and benign recurrent intrahepatic cholestasis type 1 (BRIC1), respectively. There is currently no practical method for determining PFIC1 or BRIC1 at an early disease course phase. Herein, we assessed the feasibility of developing a diagnostic method for PFIC1 and BRIC1. A nationwide Japanese survey conducted since 2015 identified 25 patients with cholestasis with ATP8B1 mutations, 15 of whom agreed to participate in the study. Patients were divided for analysis into PFIC1 (n = 10) or BRIC1 (n = 5) based on their disease course. An in vitro mutagenesis assay to evaluate pathogenicity of ATP8B1 mutations suggested that residual ATP8B1 function in the patients could be used to identify clinical course. To assess their ATP8B1 function more simply, human peripheral blood monocyte-derived macrophages (HMDMs) were prepared from each patient and elicited into a subset of alternatively activated macrophages (M2c) by interleukin-10 (IL-10). This was based on our previous finding that ATP8B1 contributes to polarization of HMDMs into M2c. Flow cytometric analysis showed that expression of M2c-related surface markers cluster of differentiation (CD)14 and CD163 were 2.3-fold and 2.1-fold lower (95% confidence interval, 2.0-2.5 for CD14 and 1.7-2.4 for CD163), respectively, in patients with IL-10-treated HMDMs from PFIC1 compared with BRIC1. Conclusion: CD14 and CD163 expression levels in IL-10-treated HMDMs may facilitate diagnosis of PFIC1 or BRIC1 in patients with ATP8B1 deficiency.
Abbreviations
-
- ATP8B1
-
- adenosine triphosphatase phospholipid transporting 8B1
-
- BRIC
-
- benign recurrent intrahepatic cholestasis
-
- CD
-
- cluster of differentiation
-
- CDC50A
-
- heterodimer complex with transmembrane protein 30A
-
- GGT
-
- gamma-glutamyltransferase
-
- HA
-
- hemagglutinin antigen
-
- HMDM
-
- human peripheral blood monocyte-derived macrophage
-
- HPBMo
-
- human peripheral blood monocytes
-
- IL-10
-
- interleukin-10
-
- M2c
-
- subset of alternatively activated macrophages
-
- MFI
-
- mean fluorescence intensity
-
- NBD-PC
-
- nitrobenzoxadiazole-labeled phosphatidylcholine
-
- PFIC
-
- progressive familial intrahepatic cholestasis
-
- WT
-
- wild type
Familial cholestatic liver disease comprises a variety of clinically and genetically heterogeneous disorders. Mutations in adenosine triphosphatase phospholipid transporting 8B1 (ATP8B1), adenosine triphosphate binding cassette subfamily B member 11 (ABCB11), tight junction protein 2 (TJP2), nuclear receptor subfamily 1 group H member 4 (NR1H4), or myosin VB (MYO5B) are responsible for autosomal recessive hereditary cholestatic diseases with normal serum gamma-glutamyltransferase (GGT) level.(1-6) Its actual prevalence remains unknown, but its estimated incidence varies between 1/50,000 and 1/100,000 births.(7)
ATP8B1 encodes a member of the P4 subfamily of P-type adenosine triphosphatases, ATP8B1, which is expressed on the apical membrane of many epithelial cells, including hepatocytes. Its correct trafficking to the plasma membrane is facilitated by a heterodimer complex with transmembrane protein 30A (CDC50A).(8) ATP8B1 translocates aminophospholipids from the outer leaflet to the inner leaflet, thereby contributing to making the hepatocanalicular membrane a rigid liquid-ordered membrane.(8, 9) In patients with ATP8B1 deficiency, the well-organized aminophospholipid asymmetry of the hepatocanalicular membrane is disrupted, leading to a decrease in the transport activity of bile salt export pump (BSEP), an ABC transporter that is localized on the hepatocanalicular membrane and mediates biliary excretion of bile salts,(10-13) and subsequently leading to the onset of severe intrahepatic cholestasis.(14) Alternatively, in patients with ATP8B1 deficiency, nuclear translocation of the farnesoid X receptor, a transcription factor that controls bile acid homeostasis, is disrupted and causes a decrease in BSEP expression at the hepatocanalicular membrane because of its transcriptional suppression.(15)
ATP8B1 deficiency causes two forms of the cholestatic diseases with normal GGT: progressive familial intrahepatic cholestasis type 1 (PFIC1) and benign recurrent intrahepatic cholestasis type 1 (BRIC1).(1) PFIC1 onset occurs around the age of 1 year, with severe unremitting cholestasis with intractable itching, jaundice, watery diarrhea, and failure to thrive, resulting in liver failure and death before adulthood.(16) No effective therapy for cholestatic liver injury of this disease is currently available.(17) While liver transplantation solves the immediate problem of liver failure in PFIC1, it is insufficient to overcome PFIC1 because of steatosis and fibrosis in the graft liver.(18, 19) BRIC1 is characterized by intermittent episodes of intrahepatic cholestasis, refractory pruritus, and jaundice. Presentation of the first attack of jaundice usually occurs at age <1-20 years.(16, 20) In most BRIC1 cases, the symptoms last for 1 to 18 months and resolve spontaneously without progressing to liver failure.(16) However, a reduction in the number and duration of the attacks is highly desirable due to intractable cholestatic pruritus. Rifampicin, cholestyramine, and nasobiliary drainage have been suggested as therapeutic options for cholestatic attacks in patients with BRIC.(21-25)
That individuals with ATP8B1 deficiency share an age of disease onset and many clinical features makes it difficult to differentiate between PFIC1 and BRIC1 early in their disease course. Given the differences in prognoses and possible treatment plans between PFIC1 and BRIC1, developing a method for correct diagnoses is of the highest priority. Genome sequence analysis, the most commonly used procedure for diagnosis of familial cholestatic liver diseases, is available to identify patients with ATP8B1 deficiency but not to distinguish between PFIC1 and BRIC1.(26) An in vitro mutagenesis assay to examine the impact of disease-causing ATP8B1 mutations on ATP8B1 function suggests that this method can predict residual ATP8B1 function in patients with ATP8B1 deficiency and may explain the difference in clinical severity between PFIC1 and BRIC1.(27) However, this procedure requires significant time and effort to identify disease-casing mutations as it involves the introduction of mutations into ATP8B1 complementary DNA (cDNA), construction of cultured cell lines with mutated ATP8B1, and evaluation of the difference in expression, localization, and activity between the wild type (WT) and mutated forms of ATP8B1. Therefore, development of a simpler faster diagnostic method to evaluate ATP8B1 function in patients with ATP8B1 deficiency is needed.
We have previously reported that ATP8B1 is expressed in human peripheral blood monocyte-derived macrophages (HMDMs) and contributes to correct polarization of HMDMs into a subset of alternatively activated macrophages (M2c)(28) that is induced by exposure to interleukin-10 (IL-10) and the function of which is related to suppression of immune responses and tissue remodeling. Decreased ATP8B1 function in IL-10-treated HMDMs markedly suppresses the expression of M2c-related surface markers cluster of differentiation (CD)14 and CD163. The present study analyzed the HMDMs from patients with ATP8B1 deficiency to evaluate it as a potential diagnostic tool for PFIC1 and BRIC1 instead of the in vitro mutagenesis assay.(27)
Patients and Methods
A detailed description is presented in the Supporting Information. All methods used standard techniques and commercially available reagents.
Ethics
The study was approved by the institutional ethics review board at the University of Tokyo and all other participating facilities and performed in accordance with the principles of the Declaration of Helsinki. Before assessment, participants or their parents (for those under age 18 years) provided signed informed consent.
Patients
The clinical diagnosis of normal-GGT PFIC or BRIC was based on the presence of unremitting or repeated intermittent hepatocellular cholestasis with intractable pruritus, jaundice with conjugated hyperbilirubinemia, elevated serum bile acid concentrations, and normal serum GGT level. Serologic, viral, or metabolic markers, imaging, and urine screening were performed to exclude other causes of cholestasis, including hepatitis B and C virus infections, inborn errors in bile-acid synthesis, and ductal origin.
A nationwide Japanese survey conducted since 2015 identified patients with normal-GGT PFIC or BRIC. In these patients, all exons and flanking intron–exon boundaries of genes responsible for neonatal/infantile intrahepatic cholestasis, including ATP8B1, were analyzed by Sanger sequencing(17, 28) and/or targeted next-generation sequencing.(29) Twenty-five patients were diagnosed with ATP8B1 deficiency because they carried mutations in both alleles of ATP8B1 or mutation in one allele of ATP8B1 without other mutations in either gene analyzed. Only 15 of these patients, excluding those who died or could not be followed, were enrolled in the study and divided for analysis into PFIC1 (n = 10) or BRIC1 (n = 5) based on their disease course (see Table 1).
| Disease | Number | Age | Sex | Cholestasis | Surgical Intervention | ATP8B1 Mutations | Predicted ATP8B1 Function* | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Onset | Forms | Exon | Nucleotide Change | AA Change | Allele (% of WT) | Patient (% of Healthy Subjects) | |||||
| PFIC1 | 1 | 3 years (Alive) | Male | 5 months | Persistent | — | 15 | c.1587_1589del | p.F529del | 0 | 0 |
| 28 | c.3628dupG | p.A1210fsX26 | 0 | ||||||||
| PFIC1 | 2 | 4 years (Alive) | Female | 7 months | Persistent | — | 5 | c.461_462insT | p.V154fsX15 | 0 | 0 |
| 19 | c.2124_2125insGAGCTACAGCTATTGAAGGC | p.K709fsX41 | 0 | ||||||||
| PFIC1 | 3 | 6 years (Alive) | Male | 2 months | Persistent | — | 19 | c.2124_2125insGAGCTACAGCTATTGAAGGC | p.K709fsX41 | 0 | 0 |
| 2-6 | exon deletion | — | 0 | ||||||||
| PFIC1 | 4 | 9 years (Alive) | Male | 2 months | Persistent | — | 25 | c.3033-34del | p.L1011fsX20 | 0 | 0 |
| N.D. | 0† | ||||||||||
| PFIC1 | 5 | 15 years (Alive) | Female | 2 months | Persistent | Biliary fistula at 1 year | 9 | c.727delC | p.L243fsX28 | 0 | 0 |
| 23 | c.2854C>T | p.R952X | 0 | ||||||||
| PFIC1 | 6 | 13 years (Alive) | Female | 4 months | Persistent | Biliary fistula at 6 years | 13 | c.1371del | p.G457fsX8 | 0 | <5 |
| 24 | c.2941G>A | p.E981K | <10‡ | ||||||||
| PFIC1 | 7 | 11 years (Alive) | Male | 3 months | Persistent | Biliary fistula at 1.5 years | 28 | c.3579_3589del | p.R1193fsX39 | 0 | 0 |
| N.D. | 0† | ||||||||||
| PFIC1 | 8 | 12 years (Dead) | Male | 3 months | Persistent | LTx at 11 years | 15 | c.1587_1589del | p.F529del | 0 | 0 |
| N.D. | 0† | ||||||||||
| PFIC1 | 9 | 7 years (Alive) | Female | 6 months | Persistent | LTx at 2 years | 10 | c.916T>C | p.C306R | <10 | <5 |
| 23 | c.2854C>T | p.R952X | 0 | ||||||||
| PFIC1 | 10 | 22 years (Alive) | Male | 7 months | Persistent | LTx at 3 years, 13 years | 13 | c.1367C>T | p.T456M | <10 | <5 |
| 21 | exon deletion | — | 0 | ||||||||
| BRIC1 | 1 | 42 years (Alive) | Female | 21 years | Intermittent attacks at 21 years, 27 years, 38 years | — | 23 | c.2854C>T | p.R952X | 0 | <5 |
| 13 | c.1408T>G | p.C470G | <10 | ||||||||
| BRIC1 | 2 | 34 years (Alive) | Female | 6 months | Intermittent attacks at 6 months, 5 years, 15 years, 17 years, 19 years, 22 years | — | 10 | c.916T>C | p.C306R | <10 | <10 |
| 22 | c.2600G>A | p.R867H | <10 | ||||||||
| BRIC1 | 3 | 7 years (Alive) | Male | 7 months | Intermittent attack at 7 months | — | 10 | c.922G>A | p.G308S | <10 | <5 |
| 28 | c.3579_3589del | p.R1193fsX39 | 0 | ||||||||
| BRIC1 | 4 | 38 years (Alive) | Male | 19 years | Intermittent attacks once a year since 19 years | — | 23 | c.2717T>C | p.I906T | <10 | <20 |
| 25 | c.3125T>C | p.L1042P | 30 | ||||||||
| BRIC1 | 5 | 7 years (Alive) | Female | 9 months | Intermittent attack at 9 months | — | 23 | c.2927C>T | p.A976V | <10 | <5+α |
| N.D. | Unknown | ||||||||||
In Vitro Mutagenesis Studies
HEK293T (CRL-11268) cells and CHO-K1 (CCL-61) cells were purchased from the American Type Culture Collection (Manassas, VA). HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific), and CHO-K1 cells were maintained in Ham’s F12 nutrient mixture (Thermo Fisher Scientific) supplemented with 10% FBS. Both cells were cultured at 37°C in 5% CO2 at 95% humidity.
The pShuttle vector containing human ATP8B1 cDNA C-terminally tagged with FLAG (pShuttle–ATP8B1-FLAG) and human CDC50A N-terminally tagged with hemagglutinin antigen (HA) (pShuttle–HA–CDC50A) and minigene vectors containing exons of interest and their flanking intronic sequences in ATP8B1 were constructed as described.(17, 28, 30) Mutations identified in patients with PFIC1 and BRIC1 were introduced into the WT form of pShuttle–ATP8B1-FLAG and minigene constructs by site-directed mutagenesis using the KOD-Plus mutagenesis kit (TOYOBO, Osaka, Japan). All constructed plasmids were verified by sequence analysis using an ABI 3730xl DNA Analyzer (Thermo Fisher Scientific) in GENEWIZ (Saitama, Japan).
HEK293T cells and CHO-K1 cells were transfected with the indicated plasmids by using PEI MAX (Polysciences, Warrington, PA) according to the manufacturer’s instructions. Forty-eight hours after transfection, the cells were subjected to in vitro experiments to analyze RNA splicing, protein expression, and flippase activity, as described.(28, 30) The primary antibodies are listed in Supporting Table S1. All other chemicals were of analytic grade.
Analysis of M2c Subsets From Patients and Control Subjects
Peripheral blood samples from patients with PFIC1 or BRIC1 and age-matched control subjects, including healthy individuals and those with pancreatitis, hepatitis B and C virus, and biliary atresia, were collected in ethylene diamine tetraacetic acid-2K-coated blood sampling tubes (Becton Dickinson, Tokyo, Japan). Human peripheral blood monocytes (HPBMo) were obtained using a monocyte enrichment cocktail (StemCell Technologies, Vancouver, Canada), seeded on Repcell dishes (CellSeed, Tokyo, Japan), and cultured at 37°C in an atmosphere of 5% CO2 in air at 95% humidity. HPBMo from more than 3 age-matched control subjects were pooled before the seeding and then employed as control cells for flow cytometric analysis. HPBMo were differentiated into HMDMs and then polarized into M2c subsets, as described.(28) The prepared cells were harvested by incubating the dishes on ice, stained with fluorochrome-labeled antibodies, and analyzed on a BD FACSAria II Cell Sorter (BD Biosciences, San Jose, CA) or BD FACSCelesta (BD Biosciences).
Statistical Analysis
Data are presented as mean ± SEM unless otherwise indicated. The differences between two variables and multiple variables were assessed at the 95% confidence levels using Student t tests and analysis of variance with Dunnett's or Bonferroni post hoc correction, respectively. Data were analyzed using Prism software (version 6; GraphPad Software, La Jolla, CA).
Results
ATP8B1 Mutation Types Identified in Patients With PFIC1 and BRIC1
Study participants included 10 patients with PFIC1 and 5 patients with BRIC1 (see Table 1). Genome sequencing identified mutations in both alleles of ATP8B1 in 11 patients (7 patients with PFIC1 and 4 patients with BRIC1). The remaining patients had mutation in one allele of ATP8B1 without other detected mutations in either gene analyzed. ATP8B1 deficiency in the other allele was supported by undetectable expression of ATP8B1 in liver and abnormal phenotypic features in IL-10-treated HMDMs, as described.(28)
Seventeen mutations identified in patients with PFIC1 were frameshift, nonsense, and exon-deletion mutations, resulting in a complete loss of function of ATP8B1, except for three missense mutations, c.916T>C (p.C306R), c.1367C>T (p.T456M), and c.2941G>A (p.E981K); further, nine mutations identified in patients with BRIC1 were comprised of seven missense mutations, one nonsense mutation, and one frameshift mutation (see Table 1). Although severe mutations were more prevalent in PFIC1 compared with BRIC1, information of mutation types was insufficient to make a diagnosis of PFIC1 or BRIC1.
Pathogenicity and Severity of Missense Mutations of ATP8B1 Identified in Patients With PFIC1 and BRIC1
To confirm that the identified missense mutations were pathogenic disease-causing mutations and not rare normal variants,(28) we evaluated the impact of these mutations on RNA splicing, protein expression, localization, and flippase activity by in vitro mutagenesis study using HEK293T cells and CHO-K1 cells (Fig. 1). In a minigene assay, which was used to evaluate the effects on premessenger RNA splicing,(30, 31) no mutation tested had an influence on the band size of products from WT ATP8B1 exons, indicating that no aberrant splicing was caused by these mutations (Fig. 1A). In contrast, most of the mutations severely decreased ATP8B1 protein expression to less than 10% of WT ATP8B1 (Fig. 1B,C). The results of the mutation, which caused more than 90% decrease in ATP8B1 expression, are represented as below quantification limit in Fig. 1C and <10% of WT in Table 1 because there was a large variation in the quantification values of ATP8B1 expression between experiments in these mutations. Three mutations, c.922G>A (p.G308S), c.2927C>T (p.A976V), and c.3125T>C (p.L1042P), which caused moderate reduction in ATP8B1 protein expression, were further analyzed to investigate the influence on localization of ATP8B1 by cell-surface biotinylation, a method used to isolate the plasma membrane fraction where ATP8B1 works to translocate aminophospholipids, such as phosphatidylcholine, from the outer leaflet to the inner leaflet.(8, 32) Cell-surface expression of ATP8B1 was decreased by more than 90% by c.922G>A (p.G308S) and c.2927C>T (p.A976V) but was only moderately affected by c.3125T>C (p.L1042P) (Fig. 1D,E). To explore the effect of c.3125T>C (p.L1042P) on intrinsic ATP8B1 function, it was further analyzed by flippase assay in which incorporation into the inner leaflet of the plasma membrane of nitrobenzoxadiazole-labeled phosphatidylcholine (NBD-PC) for 15 minutes was measured using flow cytometry.(28) c.3125T>C (p.L1042P) affected NBD-PC incorporation by ATP8B1 to the same extent as its cell-surface expression, suggesting little impact of this mutation on ATP8B1 functional activity (Fig. 1F).
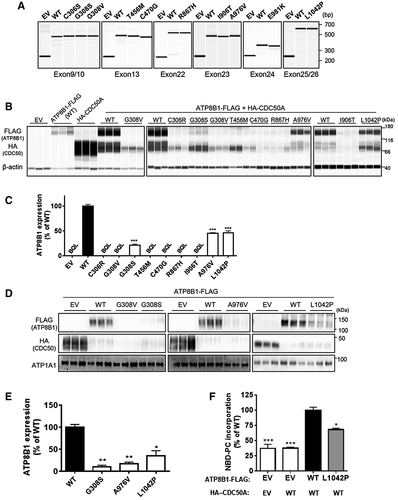
All these results suggested that the missense mutations identified from the participants, except for c.3125T>C (p.L1042P), decreased ATP8B1 function to less than 10% of WT ATP8B1.
Prediction of ATP8B1 Function in Patients With PFIC1 and BRIC1
Based on the hypothesis that ATP8B1 transcripts are generated equally from both alleles, participants’ ATP8B1 function was calculated using information of mutation types (loss of function or missense mutation) and data of pathogenicity and severity of missense mutations (Table 1; Figs. 1 and 2A). In PFIC1, it was predicted that there would be loss of function of ATP8B1 in 7 patients and less than 5% of ATP8B1 function in 3 patients relative to healthy subjects whereas there would be no patients with BRIC1 with loss of function of ATP8B1. Three patients with BRIC1 were predicted to possess more than 5% of ATP8B1 function relative to healthy subjects (Fig. 2B; Table 1). These results suggested that the remaining activity of ATP8B1 in patients with ATP8B1 deficiency was associated with their clinical courses (i.e., PFIC1 or BRIC1).

Discrimination Between PFIC1 and BRIC1 in Patients With ATP8B1 Deficiency Using IL-10-Treated HMDMs
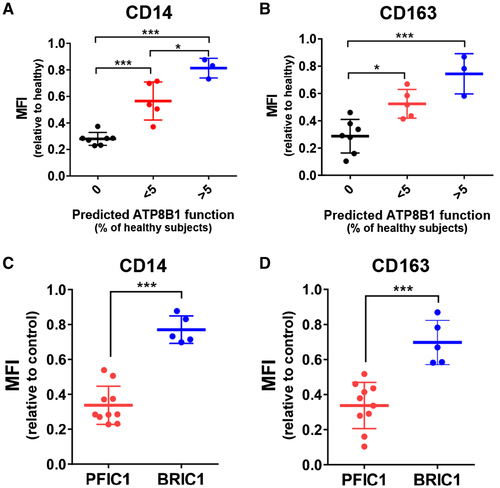
ATP8B1 contributes to correct polarization of HMDMs into M2c, a subset of alternatively activated macrophages.(28) HPBMo from patients with PFIC1 (n = 10) and BRIC1 (n = 5) were differentiated into HMDMs, and their polarization to M2c was facilitated by IL-10 stimulation. In each experiment, IL-10-treated HMDMs from control subjects were prepared from the HPBMo of at least 3 individuals to minimize interindividual variability.(28) IL-10-treated HMDMs prepared from the patients and control subjects were gated according to forward scatter/side scatter properties (Supporting Fig. S1) and analyzed to evaluate the mean fluorescence intensity (MFI) of CD14 and CD163, the surface markers of M2c.(33) The MFI value was correlated with predicted ATP8B1 function (Figs. 2B, 3A,B) and was decreased severely in PFIC1 and slightly in BRIC1. The MFIs of CD14 and CD163, calculated relative to the values in control subjects, were 2.3-fold (95% confidence interval [CI], 1.9-2.6; P < 0.001) and 2.1-fold (95% CI, 1.6-2.5; P < 0.001), respectively, lower in patients with PFIC1 than in patients with BRIC1 (Fig. 3C,D). These results suggest that residual ATP8B1 function could reflect the expression levels of CD14 and CD163 in IL-10-treated HMDMs.
Discussion
Based on genome sequence analysis and in vitro mutagenesis assay, this study of 15 Japanese patients with cholestasis with ATP8B1 deficiency first confirmed that their clinical courses depended on residual ATP8B1 function (Fig. 2), which is consistent with reports of other predominantly European patient cohorts.(26, 27) To determine their ATP8B1 function more simply, HMDMs from these patients were assessed based on our previous finding that ATP8B1 contributes to M2c polarization of HMDMs by IL-10 treatment.(28) Flow cytometric analysis provided novel evidence that expression levels of the M2c markers CD14 and CD163 in IL-10-treated HMDMs are significantly lower in PFIC1 compared with BRIC1 (Fig. 3C,D). Given that HMDMs can be prepared within 10 days from peripheral blood, which is collected less invasively, the application of these findings as a replacement for the in vitro mutagenesis assay may facilitate early diagnosis of PFIC1 and BRIC1 in patients with ATP8B1 deficiency. Although a flippase assay using fluorescent-labeled aminophospholipids may allow evaluation of intrinsic ATP8B1 function in IL-10-treated HMDMs, the flippase assay requires much more time and effort compared with evaluation of cell-surface markers by flow cytometry. The change in CD14 and CD163, a phenotypic feature resulting from ATP8B1 deficiency, is appropriate for diagnostic use of IL-10-treated HMDMs to assess ATP8B1 function.
This study suggests that the clinical course for most patients deficient in ATP8B1 (10 of 15 in this sample) is clearly determined based on their residual ATP8B1 function. Genome sequence analysis and in vitro mutagenesis assay showed that 70% of patients with PFIC1 but no patients with BRIC1 were predicted to have loss of function of ATP8B1; however, 60% of patients with BRIC1 but no patients with PFIC1 were predicted to possess more than 5% of ATP8B1 function relative to healthy subjects (Fig. 2B). These patients could also be correctly classified by phenotypic analysis of IL-10-treated HMDMs (0% vs. >5%; Fig. 3A,B). The remaining 5 patients, each of whom had <5% ATP8B1 function relative to healthy subjects, included patients with PFIC1 and those with BRIC1, indicating a limitation of the diagnostic value of in vitro mutagenesis assay with patients deficient in ATP8B1. The exogenous overexpression system may be unsuitable for predicting the impact of their mutations on ATP8B1 function. However, it is possible that this was correctly predicted using an in vitro mutagenesis assay. Other hereditary and environmental factors as well as their ATP8B1 mutations could affect these patients’ ATP8B1 function. Understanding endogenous ATP8B1 function using IL-10-treated HMDMs may facilitate diagnoses of these patients because expressions of CD14 and CD163 in 3 patients with BRIC1 and 2 patients with PFIC1 were in the top three values and the bottom two values, respectively, among 5 patients with <5% ATP8B1 function relative to healthy subjects (<5%; Fig. 3A,B). Further study with a larger patient cohort will be needed to draw definitive conclusions.
M2c macrophages preferentially and efficiently clear early apoptotic cells derived from normal homeostasis, tissue turnover, and immune responses against pathogens.(34, 35) Therefore, M2c suppresses persistent apoptosis and the accumulation of secondary necrotic cells, which are highly inflammatory because their autolysis releases cytotoxic, proinflammatory, and immunogenic molecules.(36) The anti-inflammatory actions of M2c are amplified and prolonged by their uptake of apoptotic cells and subsequent IL-10 secretion, generating a positive feedback loop of M2c homeostasis.(34, 35) Given that cholestatic liver injury is initiated by neutrophil recruitment through hepatocyte-derived chemokines,(37) abnormal M2c function could result in impaired clearance of activated and aged neutrophils, ongoing inflammation, and uncontrolled tissue damage. Expression levels of the M2c markers CD14 and CD163 in IL-10-treated HMDMs were reduced severely in PFIC1 but only slightly in BRIC1 (Fig. 3C,D), suggesting that polarization of HMDMs into M2c is severely damaged in PFIC1 but not in BRIC1. It is possible that this different function in M2c underpins the pathologic changes in liver by cholestasis in ATP8B1 deficiency in which PFIC1 but not BRIC1 progresses to cirrhosis and liver failure.(16) Inflammatory resolution by M2c function might also contribute to pathogenesis of extrahepatic manifestations related to inflammation, including recurrent pancreatitis(38) and atherosclerosis(39) because these manifestations have not been reported in patients with BRIC1. Further studies to clarify the roles of ATP8B1 in M2c and the pathophysiologic functions of M2c should facilitate our understanding of the mechanism of progressive cholestatic liver failure and the extrahepatic manifestations in PFIC1, which would allow the development of new therapies for PFIC1.
In conclusion, our study shows that expression levels of CD14 and CD163 in IL-10-treated HMDMs are significantly lower in PFIC1 than in BRIC1. The application of these findings as an early diagnostic tool in patients with ATP8B1 deficiency must be validated by future studies with larger samples than was possible in this study. If confirmed, this method may help describe prognoses and determine optimal treatment plans in patients with ATP8B1 deficiency.
Acknowledgment
We thank the patients who participated in this study and their families.