Frequent Antiviral Treatment Failures in Patients Infected With Hepatitis C Virus Genotype 4, Subtype 4r
Abstract
Hepatitis C virus (HCV) genotype 4 is highly heterogeneous. HCV subtype 4r has been suggested to be less responsive to direct-acting antiviral (DAA) drug treatment than other genotype 4 subtypes. Among 537 DAA-treated patients who experienced a virological failure (VF) in France between 2015 and 2018, 121 (22.5%) were infected with genotype 4 and 27 of them (22.3%) with subtype 4r; subtype 4r was thus over-represented as compared to its prevalence in the French general population. Population sequencing of the nonstructural protein (NS) 3, NS5A, and NS5B genes was performed in all subtype 4r patients at treatment failure and in 6 at baseline, whereas full-length HCV genome sequencing was performed in two baseline and three treatment failure samples by means of an original shotgun metagenomics method based on deep sequencing. At treatment failure, all subtype 4r patients harbored two to three dominant NS5A resistance-associated substitutions (RASs), including at least L28A/C/I/M/V and L30R. Among 13 patients exposed to sofosbuvir and an NS5A inhibitor (daclatasvir, ledipasvir, or velpatasvir), 5 (38.5%) also harbored NS5B S282C/T RASs at treatment failure. An additional patient harbored S282C/T RASs at treatment failure by deep sequencing. Prevalence of S282C/T RASs at treatment failure was significantly higher in patients infected with genotype 4r than with other genotypes, including other subtypes of genotype 4. Conclusion: The lower rates of sustained virological response in patients infected with subtype 4r are related to the frequent preexistence at treatment baseline and subsequent selection by DAA treatment of both NS5A and NS5B S282 RASs. Our study suggests that these patients should be identified and receive a triple DAA combination regimen as first-line treatment.
Abbreviations
-
- DAA
-
- direct-acting antiviral
-
- HCV
-
- hepatitis C virus
-
- NS
-
- nonstructural protein
-
- RASs
-
- resistance-associated substitutions
-
- SVR
-
- sustained virological response
-
- VF
-
- virological failure
-
- WT
-
- wild type
Since 2014, several direct-acting antiviral (DAA) drugs with potent effectiveness against hepatitis C virus (HCV) have been approved worldwide. Safe and well-tolerated combinations of these drugs yield very high rates of sustained virological response (SVR), defined as an undetectable HCV RNA 12 or 24 weeks after the end of treatment.1 SVR corresponds to a definitive cure of infection and is associated with reduced liver- and non-liver-related morbidity and mortality.2 International guidelines have been published to guide treatment decisions and management in order to optimize the results of anti-HCV therapy.1, 3 In addition, real-world studies have confirmed the safety and efficacy of HCV-DAA combination regimens.4-10 Virological failures (VFs), most often posttreatment relapses, are rare. They are generally associated with selection of viruses resistant to one or several of the drugs administered. Resistant viruses are characterized by presence of resistance-associated substitutions (RASs) in their genomes, that is, substitutions that confer reduced susceptibility to the corresponding drug or drug class. These RASs are generally present before any treatment, as naturally occurring polymorphisms carried by major or, more often, minor viral populations.11
HCV genotype 4 is highly prevalent in the Middle East and in parts of Africa.12 Its incidence and prevalence is steadily rising in Western Europe.13 Despite considerable subtype heterogeneity, SVR rates in patients with genotype 4 infections have been reported to be as high as with other genotypes.14-16 Indeed, in a recent compilation of 18 clinical trials with various HCV treatment regimens submitted to the U.S. Food and Drug Administration between 2014 and 2017, only 12 of 573 patients infected with genotype 4a (2.1%) experienced VF. Relapse occurred in 10 patients, including 6 infected with subtype 4d, 2 with subtype 4r, 1 with subtype 4b, and 1 with an unknown genotype 4 subtype. On-treatment breakthrough occurred in 2 participants, 1 each with subtype 4a and 4d. Overall, 2 of 12 patients infected with subtype 4r failed to achieve SVR.17 A recent prospective study conducted in Rwanda in adult patients infected with HCV genotype 4 treated with a one-pill combination of the nucleotide analogue inhibitor of the HCV RNA-dependent RNA polymerase sofosbuvir and the nonstructural (NS) 5A protein inhibitor, ledipasvir, identified one of the genotype 4 subtypes, subtype 4r, as less responsive than other subtypes (SVR rates, 54% for subtype 4r vs. 93% for 4k; 90% for 4q; 100% for 4v; and 87% for other subtypes).18
Subtype 4r is common in many African countries, including Uganda, the Democratic Republic of Congo,12 the Central African Republic,19 or Gabon.20 It is present, but rarely found, in high-income countries, including Europe and the United States. In France, 12.4% (164 of 1,322) of all patients with chronic hepatitis C (CHC) prospectively included between 2010 and 2014 in a nation-wide, multicenter cohort study (CO22 HEPATHER) were infected with genotype 4.13 In the pooled PEARL-I and AGATE-I studies, clinical trials including patients treated with ombitasvir/paritaprevir/ritonavir, only 1 of 101 French genotype 4–infected patients was infected with subtype 4r.21 In the pooled GS-US-337-1119 and ASTRAL-1 trials, which included patients from Europe and the United States treated with sofosbuvir/ledipasvir and sofosbuvir/velpatasvir, respectively, 5 of 160 genotype 4–infected patients (i.e., 3.1%) were infected with subtype 4r.22
The aim of this study was to assess the proportion of subtype 4r infections in a population of patients treated for their HCV infection in France who failed to achieve SVR and to virologically characterize their treatment failures.
Patients and Methods
Population Sequencing of Three Genomic Regions
The French National Reference Center for Viral Hepatitis B, C and Delta performs HCV genotypic resistance testing for patients with CHC who failed to achieve SVR across the country. In each patient referred, three genomic regions targeted by the HCV DAAs, including the NS3 protease, NS5A protein, and NS5B polymerase coding regions, have been analyzed by means of population sequencing (sensitivity, approximately 15%) at treatment failure and, when available, at baseline, as recently described.23 Briefly, HCV RNA was extracted with the QIASymphony DSP Virus/Pathogen kit on a QIASymphony device (Qiagen GmbH, Hilden, Germany), according to the manufacturer's instructions. Complementary DNA synthesis was performed with the OneStep RT-PCR kit (Qiagen GmbH) with sets of primers adapted to the viral regions targeted.22 Nested PCR was then performed with common primers and, when necessary, genotype-specific primers. PCR products were purified with Amicon Ultra-0.5 mL Centrifugal Filters (EMD Millipore, Darmstadt, Germany) and sequenced with the BigDye Terminator Cycle Sequencing Kit v3.1 on an ABI Prism 3730 DNA Analyzer (Applied Biosystems, Foster City, CA).
Phylogenetic analysis was carried out by means of the Phylogeny Inference Package (PHYLIP; version 3.695), using genotype 1-7 subtype reference sequences available in GenBank. Nucleotide sequences (position 724-1,009 according to the H77-1a prototype strain) were aligned with reference sequences using CLUSTAL W.24 Phylogenetic relationships were deduced by means of DNADIST-NEIGHBOR from PHYLIP. For neighbor-joining analysis, a Kimura two-parameter distance matrix with a transition/transversion ratio of 2.0 was used.25 Phylogenetic trees were plotted with FigTree v1.4.3.26 Their robustness was assessed by bootstrap analysis of 1,000 replicates by means of the SEQBOOT program from PHYLIP.
Full-Length HCV Genome Sequence Analysis by Means of Shotgun Metagenomics using Deep Sequencing
Full-length HCV genome sequences were generated by means of an in-house technique based on shotgun metagenomics using deep sequencing. Briefly, after pretreatment with a combination of mechanical and enzymatic fragmentations, RNA was extracted by means of the DNA midi kit on a QiaSymphony device (Qiagen GmbH). Quality (Tape Station; Agilent, Santa Clara, CA) and quantitative yield (Quant-it; Thermo Fisher Scientific, Waltham, MA) of extracted nucleic acids were verified. Sequencing used the TruSeq Total RNA kit (Illumina, San Diego, CA) after automated library preparation on a MicroLab StarLet device (Hamilton Robotics, Reno, NV). Twelve-sample multiplexing was performed to allow for minor variant detection. Sequencing was run on a NextSeq500 device with the sequencing kit, High Output 2*150 (Illumina). Sequences were analyzed by means of in-house software (MetaMIC), developed for bioanalysis of microbiological big data generated by metagenomics. MetaMIC uses different modules for demultiplexing, quality check, dusting, pair end-long sequence identification using several different databases, genotyping and mapping of viral sequences with the best reference sequence, variant calling, and rates of nucleotide or amino acid changes estimation. The cutoff for detection of minority variants was set up at 1%, as is usually the case for deep sequencing studies of viral quasi-species. Full-length HCV genome sequences have been submitted to GenBank.
Ethical Aspects
The study was conducted in accord with the Declaration of Helsinki and French law for biomedical research and was approved by our institution's institutional review board.
Results
Patients Infected with Subtype 4r
Between 2015 and 2018, 537 patients treated with a DAA-containing regimen who experienced a VF were analyzed. One hundred twenty-one patients (22.5%) were infected with genotype 4; among them, 27 (22.3%) were infected with subtype 4r, based on population sequencing and phylogenetic analysis of the NS5B region. A pretreatment sample was available for 6 of these 27 patients and was also analyzed. Characteristics of the 27 patients infected with subtype 4r are shown in Table 1. All of them were of African origin. In addition, one patient (P1) had previously failed the combination of sofosbuvir and ledipasvir for 12 weeks. All patients relapsed after receiving the full course of therapy.
| Patient | Age (Years) | Sex | DAA-Containing Regimen | Sample | RASs at Baseline and/or Treatment Failure | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NS3 Protease RAS Positions | NS5A RAS Positions | NS5B RAS Positions | |||||||||||||||||
| 36 | 41 | 56 | 80 | 155 | 156 | 168 | 24 | 28 | 30 | 31 | 58 | 93 | 282 | 321 | |||||
| P1 | 67 | F | SOF/VEL + RBV 12w | Failure | L | Q | Y | Q | R | A | D | K | V | R | M | P | H | S | V |
| P2* | 67 | F | SOF/LDV 12w | Baseline | L | Q | Y | Q | R | A | D | K | V | R | M | P | Y | S | V |
| Failure | L | Q | Y | Q | R | A | D | K | V | R | M | P | Y | T | V | ||||
| P3 | 42 | M | SOF/LDV 12w | Failure | L | Q | Y | Q | R | A | D | K | T | H | L | P | Y | S | I |
| P4 | 66 | F | SOF/LDV 12w | Failure | L | Q | Y | Q | R | A | D | K | V | R | M | P | Y | S | I |
| P5 | 64 | F | SOF/LDV 12w | Failure | NA | NA | NA | NA | NA | NA | NA | K | V | R | M | P | Y | S | I |
| P6 | 55 | M | SOF/LDV 12w | Baseline | L | Q | Y | Q | R | A | D | K | V | R | L | T | Y | S | V |
| Failure | L | Q | Y | Q | R | A | D | K | V | R | M | T | Y | T | V | ||||
| P7 | 59 | M | SOF/LDV 12w | Failure | L | Q | Y | Q | R | A | D | K | M | R | M | P | Y | C/T | V |
| P8 | 55 | F | SOF/LDV 12w | Failure | NA | NA | NA | NA | NA | NA | NA | K | I | R | V | P | Y | S | I |
|
P9 |
52 | M | SOF/LDV 12w | Baseline | NA | NA | NA | NA | NA | NA | NA | K | V | R | L | P | Y | S | V |
| Failure | L | Q | Y | Q | R | A | E | K | V | R | V | P | Y | S | V | ||||
| P10 | 49 | F | SOF/LDV 12w | Failure | L | Q | Y | Q | R | A | D | K | V | R | M | P | Y | C/T | V |
| P11* | 56 | F | SOF/LDV 24w | Baseline | L | Q | Y | Q | R | A | D | K | V | R | L | P | Y | S | V |
| Failure | L | Q | Y | Q | R | A | D | K | V | R | M | P | Y | T | V | ||||
| P12* | 68 | M | SOF + DCV 12w | Baseline | L | Q | Y | Q | R | A | D | K | V | R | M | P | Y | S | V |
| Failure | L | Q | Y | Q | R | A | D | K | V | S | M | P | Y | S | V | ||||
| P13 | 40 | M | SOF + DCV 12w | Failure | L | Q | Y | Q | R | A | D | K | M | R | M | P | Y | S | I |
| P14 | 63 | F | OBV/PTV/r 12w | Failure | L | Q | H | Q | R | A | V | K | C | R | L | P | Y | S | V |
| P15 | 56 | M | OBV/PTV/r 12w | Failure | L | Q | Y | Q | R | A | D | K | V | R | L | P | Y | S | V |
| P16 | 55 | F | OBV/PTV/r 12w | Baseline | NA | NA | NA | NA | NA | NA | NA | K | V | R | L | P | Y | S | V |
| Failure | NA | NA | NA | NA | NA | NA | NA | K | V | R | L | S | Y | S | V | ||||
| P17 | 60 | F | OBV/PTV/r 12w | Failure | L | Q | Y | Q | R | A | D | K | V | R | L | G | Y | S | V |
| P18 | 55 | M | OBV/PTV/r 12w | Failure | L | Q | Y | Q | R | A | D | K | V | R | L | P | Y | S | I |
| P19 | 51 | F | OBV/PTV/r 12w | Failure | L | Q | Y | Q | R | A | V | K | V | R | L | P | Y | S | V |
| P20 | 58 | M | OBV/PTV/r 12w | Failure | L | Q | Y | Q | R | A | V | K | V | R | L | P | Y | S | I |
| P21 | 61 | M | OBV/PTV/r 12w | Failure | L | Q | Y | Q | R | A | A | K | M | Q | L | P | Y | S | I |
| P22 | 61 | F | OBV/PTV/r+RBV12w | Failure | NA | NA | NA | NA | NA | NA | NA | K | A | R | L | P | Y | S | V |
| P23 | 62 | M | GZR/EBR 12w | Failure | L | Q | H | Q | R | A | V | NA | NA | NA | NA | NA | NA | S | V |
| P24 | 80 | F | GZR/EBR 8w | Failure | L | Q | Y | Q | R | A | D | K | I | R | M | S | S | S | V |
| P25 | 55 | F | SOF + SIM 12w | Failure | L | Q | Y | R | R | A | E | K | V | R | L | P | Y | S | V |
| P26 | 85 | F | SOF + SIM 12w | Failure | NA | NA | NA | NA | NA | NA | NA | K | V | R | L | P | Y | S | V |
| P27 | 76 | F | SOF + SIM 12w | Failure | L | Q | Y | Q | R | A | E | K | V | R | L | P | Y | S | V |
- RASs, that is, substitutions associated with reduced susceptibility to the corresponding drug(s), are shown in bold.
- a Indicates patients for whom full-length HCV genome sequences were analyzed by means of shotgun metagenomics using deep sequencing. Abbreviations: SOF, sofosbuvir; VEL, velpatasvir; LDV, ledipasvir; DCV, daclatasvir; OBV, ombitasvir; PTV, paritaprevir; r, ritonavir; RBV, ribavirin; GZR, grazoprevir; EBR, elbasvir; NA, not analyzed.
Baseline Sequences in Patients Infected with Subtype 4r (Population Sequencing)
At treatment baseline, NS5A polymorphisms known to be associated with reduced susceptibility to NS5A inhibitors were present in the 6 patients with available baseline sample (Table 1). Two to three NS5A polymorphisms were present as dominant species by population sequencing. The most common polymorphism was the double L28V + L30R substitution, using the genotype 4a sequence as a reference. In addition, the NS3 protease polymorphism, D168E, was present in the dominant viral population in 1 patient who had never been previously exposed to an NS3 protease inhibitor. Other key amino acid positions in the NS3 protease coding region were identical to those in the genotype 4a consensus sequence, including L36, Y56, Q80, R155, and A156. No NS5B polymerase polymorphisms were detected at baseline in the 6 patients infected with genotype 4r.
Sequences at Treatment Failure in Patients Infected with Subtype 4r (Population Sequencing)
Eleven of the 27 patients with treatment failure infected with subtype 4r had been exposed to both an NS3 protease and an NS5A inhibitor. Sequence analysis at failure was impossible for technical reasons in 1 of them. The remaining patients harbored a combination of two dominant NS5A RASs at failure, including L28A/C/I/M/V and L30R. NS3 protease RASs were also found in 5 patients (56%; Table 1).
The 13 patients exposed to sofosbuvir and an NS5A inhibitor (daclatasvir, ledipasvir, or velpatasvir) harbored viruses highly resistant to NS5A inhibitors at treatment failure, including 8 (62%) carrying the L28V + L30R combination (Table 1). Ten patients (77%) also harbored an L31M/V RAS at treatment failure. This substitution has been shown to be the most frequently associated with VF in patients infected with genotype 4 treated with daclatasvir or ledipasvir.22, 27 In addition, the triple RAS combination, L28V + L30R + L31M, has been shown to profoundly reduce the susceptibility of a genotype 4 replicon to sofosbuvir and ledipasvir (fold-change, ≈100).27 The Y93H RAS was found in only 1 patient at treatment failure.
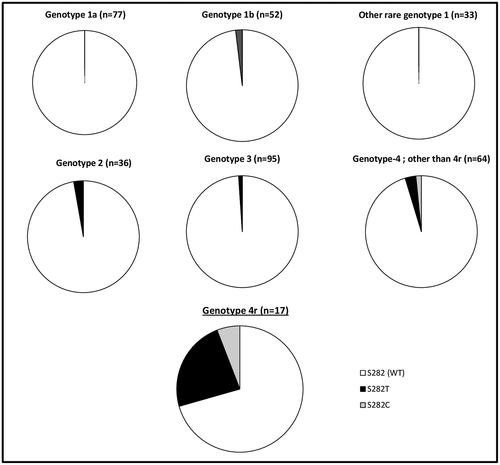
A dominant NS5B S282T substitution was detected at treatment failure in 5 of the 13 patients (38.5%) exposed to sofosbuvir and an NS5A inhibitor, alone or associated with a minority S282C population (Table 1). This substitution confers reduced susceptibility to sofosbuvir in vitro and has been exceptionally and very transiently selected by sofosbuvir-based regimens failing to achieve SVR. We retrieved our database of 381 patients infected with all genotypes who failed to achieve SVR after a sofosbuvir-containing treatment. S282C/T RAS was significantly more often found at treatment failure in patients infected with genotype 4r than in those infected with other genotypes (Fig. 1), including other subtypes of genotype 4 (P < 0.01).
Full-Length HCV Genome Sequences at Baseline and Treatment Failure in Patients Infected with Subtype 4r (Shotgun Metagenomic by Means of Deep Sequencing)
An original in-house method based on shotgun metagenomics analysis using deep sequencing and the METAMIC software was used to characterize full-length HCV genome sequences in 2 patients infected with genotype 4r at baseline and treatment failure (patients P11 and P12 in Table 1) and in an additional patient at treatment failure (patient P2 in Table 1; baseline sample not available). Deep sequencing offered two opportunities: increased sensitivity for detection of minority viral populations and full-length sequence analysis for exploration of genome regions not directly targeted by the administered DAAs.
Figure 2A shows the depth of deep sequencing along the full-length HCV genome sequence (i.e., the exact number of times that a base in the reference was covered by a high-quality aligned read from the sequencing experiment28) in the five samples tested. No particular amino acid changes were observed between baseline and treatment failure in regions other than those targeted by the HCV DAAs in the patients studied (data not shown).
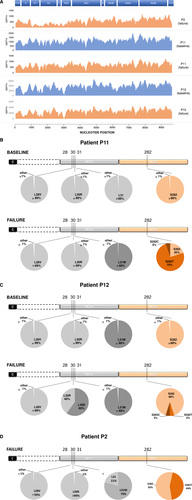
Figure 2B,C,D shows the proportions of RASs, as detected by deep sequencing, in the NS5A and NS5B regions in patients P11, P12, and P2, respectively. In patients P11 and P12, baseline NS5A and NS5B RAS profiles did not differ between deep sequencing analysis (Fig. 2B,C) and population sequencing (Table 1). Both patients harbored a wild-type (WT) S282 population representing more than 99% of the viral quasi-species in the NS5B region (Fig. 2B,C). The number of reads at NS5B position S282 were 1,168 and 4,066 in patients P11 and P12, respectively, and none of them harbored another amino acid at this position, indicating that the S282C/T RASs that were subsequently selected in these patients preexisted at extremely low levels, below the capacity of detection of highly sensitive technologies, such as the deep sequencing method used in this work. The S282 population was associated at baseline with NS5A RASs conferring high-level resistance to NS5A inhibitors, including a >99% L28V + L30R population in patient P11 and a >99% L28V + L30R + L31M population in patient P12 (Fig. 2B,C).
In patient P11 at treatment failure, population and deep sequencing reported the same >99% L28V + L30R + L31M NS5A RAS profile. In contrast, this patient was found to harbor 20% of the WT S282 population in the NS5B region, which was not detected by population sequencing. WT S282 variants were mixed with 75% of S282T and 5% of S282C, both detected by population sequencing (Fig. 2B). There were two discrepancies between population sequencing and deep sequencing in patient P12 at treatment failure: The 40% NS5A L30R RAS population had not been detected by population sequencing; similarly, the 5% and 5% sofosbuvir-resistant NS5B S282C and S282T populations, respectively, were detected only by deep sequencing (Fig. 2C). In patient P2 at treatment failure, the sofosbuvir-resistant S282T RAS was detected as present as 44% of the viral quasi-species by deep sequencing, whereas the remaining 56% of WT S282 had not been detected by population sequencing (Fig. 2D). In addition, in patient 2, the WT L31 was detected in 21% of viral variants by deep sequencing, whereas only L31M had been reported by population sequencing (Fig. 2D). These results demonstrate the key role played by the selection of RASs at position S282 of the NS5B region that confer reduced susceptibility of sofosbuvir in treatment failure in the 3 patients analyzed by means of shotgun metagenomics based on deep sequencing.
Post-Treatment Failure Retreatment Outcomes
Eight patients have been retreated in the Department of Hepatology of our center, with various regimens combining two DAAs (4 cases) or three DAAs (4 cases) with or without ribavirin for 12 or 24 weeks. SVR was achieved in 7 cases whereas treatment is still ongoing in the remaining case (Table 2). Interestingly, 3 patients with subtype 4r infection achieved SVR with glecaprevir/pibrentasvir administered for 12 weeks.
| Patient | Retreatment Regimen | Virological Response |
|---|---|---|
| P1 | No retreatment | Not applicable |
| P2 | Sofosbuvir/velpatasvir/voxilaprevir for 12 weeks | SVR12 |
| P5 | Sofosbuvir + glecaprevir/pibrentasvir for 12 weeks | SVR12 |
| P9 | Glecaprevir/pibrentasvir for 12 weeks | SVR12 |
| P11 | Glecaprevir/pibrentasvir for 12 weeks | SVR12 |
| P12 | Sofosbuvir + grazoprevir/elbasvir + ribavirin for 24 weeks | SVR12 |
| P15 | Glecaprevir/pibrentasvir for 12 weeks | SVR12 |
| P16 | Sofosbuvir/velpatasvir/voxilaprevir for 12 weeks | Ongoing |
| P25 | Sofosbuvir + daclatasvir + ribavirin for 24 weeks | SVR12 |
- Abbreviation: SVR12, SVR at 12 weeks posttreatment.
Discussion
Our data, showing a large over-representation of patients infected with HCV subtype 4r among those who failed to achieve SVR after DAA-based therapy in France, as compared to the very low prevalence of this subtype in the country,13, 15, 21, 29 indicate that HCV subtype 4r is difficult to cure with currently available antiviral drug regimens. These results are in keeping with a Rwandan study showing a 54% SVR rate in patients infected with subtype 4r treated with sofosbuvir and ledipasvir, significantly lower than that with other genotype 4 subtypes (87%-100%).18 In another study including patients treated with sofosbuvir and ledipasvir, 2 of 3 patients infected with subtype 4r failed, whereas the SVR rate was 100% in 25 subtype 4a and 10 subtype 4d patients. At treatment failure, both patients harbored a triple RAS profile in the NS5A region (L28M/V + L30R + L31M), and 1 patient also selected the NS5B S282T RAS.22 Importantly, our patients were treated with various first- and second-wave anti-HCV drug regimens, suggesting that subtype 4r is difficult to cure, regardless of the HCV regimen used.
Our extensive resistance analyses of 27 genotype 4r–infected patients at treatment failure and of 6 at baseline provide a rational explanation for the frequency of treatment failures in this particular HCV subtype. (1) Multiple NS5A RASs conferring substantially reduced susceptibility to NS5A inhibitors appear to be frequently present before treatment in these patients: Indeed, such polymorphisms were found at detectable levels in all patients tested at baseline, and multiple NS5A RASs conferring high-level resistance were selected in all patients who failed. (2) Fit viral populations harboring S282C/T RASs appear to be frequently present in patients infected with genotype 4r, probably as minority populations given that they were undetectable at baseline (even when using deep sequencing analysis), but often selected at treatment failure in our study. Given the transient nature of these variants, it is likely that more patients selected them, but they had already been replaced by WT virus at the time of resistance testing. This hypothesis is reinforced by our finding that all 3 patients tested with deep sequencing at treatment failure harbored S282C/T RASs in various proportions. Together, these features explain the reduced susceptibility of subtype 4r to both sofosbuvir and NS5A inhibitors, explaining the poor response to different regimens containing one or two of these compounds. Our results emphasize the key role played by preexisting minority S282 RASs in patients infected with subtype 4r and their selection during sofosbuvir administration, which reduces the effectiveness of sofosbuvir on variants harboring preexisting NS5A RASs, such as those observed with other HCV genotype 4 subtypes.
The lower rates of SVR in patients infected with subtype 4r are important to know, in particular for African countries in the process of implementing new DAA-based regimens (often based on generic DAAs) and in which this genotype is not rare (up to 35%).12, 19, 20, 30 Our data are also important for European countries where many HCV-infected patients originating from these areas live and will be treated in the future.
Our results raise the question of subtype 4r identification in clinical practice. Presently, the most frequently used standardized commercial HCV genotyping assays worldwide are the reverse hybridization-based line probe assay, VERSANT HCV Genotype 2.0 Assay (Siemens Healthcare, Erlangen, Germany), which targets both the 5’ untranslated and the core-coding regions of the HCV genome, and the real-time PCR-based assay, Abbott RealTime HCV Genotype II (Abbott Molecular, Chicago, IL), which targets the NS5B RNA-dependent, RNA polymerase–coding region of the HCV genome. Despite the broad variability of genotype 4, none of these assays is able to identify genotype 4 subtypes.31 Our results suggest that, given the lower likelihood of an SVR with generally recommended first-line HCV therapies, probes specific for HCV subtype 4r should be introduced in these two assays in order for them to recognize this subset of patients. Meanwhile, genotype 4r can be identified in settings using in-house population sequencing of the NS5B region followed by phylogenetic analysis, provided that reference subtype 4r sequences have been implemented in the database. A standardized assay based on deep sequencing of a portion of the NS5B region (nucleotide positions 990-1,677 according to the H77 genotype 1a prototype strain), followed by automated classification by means of nucleotide sequence homology and phylogenetic analysis, has been recently developed. We showed that this assay identifies multiple HCV subtypes, including subtype 4r.32 Thus, technical options exist and others should be developed for the identification of HCV subtype 4r, which has clinical implications in settings where patients of African origin represent a substantial population.
Whether HCV resistance testing should be recommended before first-line therapy in patients of African origin or in those infected with genotype 4 is debatable. The current trend, illustrated by recent international society guidelines, is to use pangenotypic regimens as first-line therapies without previous resistance testing.1, 3 The simple identification of subtype 4r, as discussed above, would provide sufficient information to reinforce treatment in patients infected with this subtype. However, HCV resistance testing can be used confidently in settings that have easy access to a reliable, quality-controlled assay, as recently reported.33-35 Classically, three coding regions are analyzed, including NS3 protease, NS5A, and NS5B RNA polymerase, whereas in-house databases or free-access Internet tools are used for sequence interpretation. It is, however, important to note that NS5A RASs preexisting at treatment baseline were easily found by population or deep sequencing in our patients, but this was not the case for RASs at NS5B position S282, which appear to be the main drivers of sofosbuvir-based treatment failures in these patients and were present as minor, undetectable populations at baseline in our study (including when using deep sequencing). Thus, accurate identification of subtype 4r appears to be more useful to tailor therapy than pretreatment HCV resistance assessment.
Our study shows that patients infected with genotype 4r may fail many of the recommended first-line HCV treatment regimens, including sofosbuvir plus simeprevir, sofosbuvir plus daclatasvir, sofosbuvir/ledipasvir, sofosbuvir/velpatasvir, ombitasvir/paritaprevir/ritonavir, and grazoprevir/elbasvir, a result in keeping with the reduced susceptibility to these NS5A inhibitors conferred by RASs at positions 28, 30, and/or 31. Thus, our results suggest that patients identified with subtype 4r should receive a triple DAA combination as first-line treatment (sofosbuvir/velpatasvir/voxilaprevir, sofosbuvir plus glecaprevir/pibrentasvir, or sofosbuvir plus grazoprevir/elbasvir if none of the two former combinations is available). Interestingly, 4 of our patients achieved SVR after retreatment with one of these triple DAA combination regimens. However, it is interesting to note that 3 other patients achieved SVR after retreatment with glecaprevir/pibrentasvir for 12 weeks, possibly reflecting the rarity of the NS5A Y93H RAS in patients infected with subtype 4r, pibrentasvir having substantially greater effectiveness than other NS5A inhibitors against RASs at positions 28, 30, and 31. Future treatment recommendations will rely on studies validating these treatment regimens in subtype 4r–infected patients and on the ability of testing facilities to accurately determine the genotype 4 subtype in regions where this subtype is present.
Potential conflict of interest
Dr. Ruiz consults for AbbVie. Dr. Rodriguez received grants from Gilead, Vela, and Illumina, has served as an advisor for AbbVie, Illumina, and Vela diagnostics. Dr. Pawlotsky consults for and received grants from Abbott, AbbVie, and Gilead. He consults for Abbvie, Gilead, Merck, and Abbott. Dr. Hezode advises for and is on the speakers bureau for AbbVie, Bristol-Myers Squibb, Gilead, and Merck. Dr. Fourati has served as an advisor for Abbvie. Dr. Hézode has served as a Speaker and adviser for Abbvie, BMS, Gilead, and Merck. Dr. Chevaliez has received research grants from Gilead and French Ministry of Health, and also served as an advisor for Abbott, Abbvie, Gilead, Hologic, and Cepheid.




