Oxidatively Modified Protein-Disulfide Isomerase–Associated 3 Promotes Dyskerin Pseudouridine Synthase 1–Mediated Malignancy and Survival of Hepatocellular Carcinoma Cells
Potential conflict of interest: Nothing to report.
Supported by the Korea Healthcare Technology Research and Development Project of the Ministry of Health and Welfare of the Republic of Korea (HI14C2094) and the National Research Foundation of Korea grant funded by the Korean government (NRF-2016R1A2B4008083).
Abstract
Dyskerin pseudouridine synthase 1 (DKC1) is a conserved gene encoding the RNA-binding protein dyskerin, which is an essential component of the telomerase holoenzyme. DKC1 up-regulation is frequently observed in many different human cancers including hepatocellular carcinoma (HCC); however, its regulatory mechanisms remain unclear. Thus, we investigated the regulatory mechanism of DKC1 in HCC progression. We found that protein-disulfide isomerase-associated 3 (PDIA3) interacted with the DKC1 regulatory DNA in HCC cells but not in HCC cells with elevated reactive oxygen species (ROS) levels, using liquid chromatographic–tandem mass spectrometric analysis after isolating the DKC1 regulatory region binding proteins. PDIA3 repressed DKC1 expression in HCC cells by recognizing the G-quadruplex DNA at the DKC1 location. However, oxidative modification of PDIA3 induced by ROS redistributed this protein into the cytosolic regions, which stimulated DKC1 expression. We also identified Met338 in PDIA3 as the oxidatively modified residue and validated the effect of oxidative modification using an ectopic expression system, a clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 knock-in system, and a xenograft mouse model. We observed that oxidatively modified PDIA3 promoted DKC1-mediated malignancy and survival of HCC cells in vitro and in vivo. HCC tissues showed a positive association with ROS, cytoplasmic PDIA3, and nuclear DKC1 levels. HCC patients with high PDIA3 protein and DKC1 mRNA levels also displayed reduced recurrence-free survival rates. Cumulatively, the results showed that cytoplasmic PDIA3 activity could be essential in raising DKC1 expression in HCC progression and predicting poor prognoses in HCC patients. Conclusion: Our study indicates that the elevated ROS levels in HCC modulate cytoplasmic PDIA3 levels, resulting in HCC cell survival through DKC1 up-regulation.
Abbreviations
-
- ChIP
-
- chromatin immunoprecipitation
-
- CRISPR
-
- clustered regularly interspaced short palindromic repeats
-
- DKC1
-
- dyskerin pseudouridine synthase 1
-
- G4
-
- G-quadruplex
-
- HCC
-
- hepatocellular carcinoma
-
- LC/MS-MS
-
- liquid chromatography–tandem mass spectrometry
-
- M338
-
- methionine 338
-
- NLS
-
- nuclear localization signal
-
- PDIA3
-
- protein-disulfide isomerase-associated 3
-
- ROS
-
- reactive oxygen species
-
- siRNA
-
- short interfering RNA
-
- WT
-
- wild type
Dyskerin pseudouridine synthase 1 (DKC1) is a conserved gene encoding the RNA-binding protein dyskerin, which is an essential component of telomerase holoenzyme.1 Telomerase holoenzyme functions in a major pathway for telomere maintenance, and the majority of cancers overcome the telomere shortening–mediated crisis through telomerase reactivation.1 Several lines of evidence suggest a telomerase-independent DKC1 function because DKC1 depletion inhibits the proliferation of tumor cells in which telomeres are maintained by a telomerase-independent telomere maintenance mechanism termed “alternative lengthening of telomeres.”2, 3 DKC1 mediates pseudouridylation of spliceosomal mRNA, which is critical during precursor mRNA maturation, and pseudouridylation of ribosomal RNA (rRNA), which is a central process during ribosome biogenesis.1, 3 In particular, dysregulated ribosome biogenesis can lead to diseases such as dyskeratosis congenita and cancers by inducing changes in levels of specific proteins.1, 4 Reports have shown that DKC1 expression levels increased in malignant tumors compared to those in nontumors.1, 2, 4
Numerous studies involving DKC1 have been reported, including studies on the characterization of DKC1 deficiency–mediated diseases, roles of DKC1 in rRNA and mRNA processing, actions of DKC1 in telomere restoration, and observation of highly expressed DKC1 in various human cancers.1, 2, 4 Because of the discovery of transcriptional factors regulating the human DKC1 promoter, only a small number of papers have focused on DKC1 transcription.2, 5 N-Myc and c-Myc target the DKC1 promoter and increase DKC1 mRNA levels in neuroblastomas.2 Sp1 and Sp3 bind the promoter of DKC1, where Sp1 acts as a transcriptional activator and Sp3 acts as a transcriptional inhibitor in Hela and HEK293 cells.5 Liver hepatocellular carcinoma (HCC) tissues express high levels of dyskerin, but the regulatory mechanisms of DKC1 transcription in HCC remain unclear.6
The liver is a highly metabolic organ, and reactive oxygen species (ROS) as a by-product of metabolism are mostly generated in the liver.7 ROS encompasses singlet oxygen (O2), hydrogen peroxide (H2O2), superoxide anion (O2–), and the hydroxyl radical (OH•).8 ROS are involved in cancer development through regulation of gene transcription by epigenetic modification of regulatory DNA regions.9, 10 Promoter methylation by ROS has been reported for tumor suppressor genes and for genes encoding antioxidant enzymes and their regulators.9, 11 ROS also influence cancer development by the recruitment and modulation of transcription factors.9, 12 ROS increase nuclear localization and DNA-binding activity of transcription factors such as nuclear factor kappa B and activating protein-1, which control tumor cell proliferation and survival.13, 14 ROS induce phosphorylation of ataxia-telangiectasia mutated (ATM) protein kinase, tumor suppressor p53, and checkpoint kinase 2, which are essential components in the DNA damage response.15 ATM oxidized by H2O2 forms disulfide dimers and becomes an active kinase, resulting in increased binding of and phosphorylation at p53.15 These observations indicate the role of ROS as regulators of protein function through oxidative modification. Recent papers have reported that a DKC1 deficiency triggers oxidative stress irrespective of telomere maintenance,16 whereas another DKC1 isoform allows cells to acquire tolerance to increased ROS levels.17 This explains the association between the DKC1 protein and the ROS-related metabolism. However, the causal relationship between the DKC1 protein and the ROS-mediated oxidative modification remains unknown. In this study, we show that H2O2 increased DKC1 expression through oxidative modification of protein-disulfide isomerase-associated 3 (PDIA3, known as Erp57). In this process, the function of PDIA3 as a transcriptional repressor of DKC1 was inhibited, while H2O2 increased PDIA3-mediated reductase activity, tumor cell survival, and invasive ability. These features of PDIA3 were dependent on oxidative modification of methionine 338 (M338). Therefore, elevated H2O2 levels modulated PDIA3 action in tumor cell metabolism, including DKC1 transcriptional regulation and tumor cell proliferation, leading to malignant liver tumor progression.
Materials and Methods
IDENTIFICATION OF THE OXIDATION-SENSITIVE RESIDUES OF PDIA3
The recombinant human PDIA3 protein was dialyzed against 20 mM Tris (pH 7.4). PDIA3 protein (0.5 μg) was mixed with H2O2 (final concentrations of 0, 10, 1,000, and 10,000 μM). Additional details are described in the Supporting Information.
G-QUADRUPLEX MOTIF PREDICTION AND GEL RETARDATION ASSAY
We predicted the G-quadruplex (G4) DNA structure of DKC1 using QuadBase (http://quadbase.igib.res.in/) and QGRS Mapper (http://bioinformatics.ramapo.edu/QGRS/). Additional details are described in the Supporting Information.
HUMAN SAMPLES
The experiments using human tissues were approved by the Seoul National University institutional review board (E1308/001-035). Additional details are described in the Supporting Information.
ESTABLISHMENT OF M338L HETEROZYGOUS KNOCK-IN HCC CELLS USING CLUSTERED REGULARLY INTERSPACED SHORT PALINDROMIC REPEATS (CRISPR)/CRISPR-ASSOCIATED–DERIVED RNA-GUIDED ENGINEERED NUCLEASES
The design and validation of RNA-guided engineered nucleases and the establishment of heterozygous human PDIA3 knock-in cells were performed by ToolGen (Korea). Additional details are described in the Supporting Information.
STATISTICAL ANALYSIS
The P values shown in the HCC cell data relative to the mRNA expression levels, chromatin immunoprecipitation (ChIP), promoter activity, fluorescence intensity, disulfide isomerase activity, caspase 3/7 activity, cell viability, and invasion were calculated by a standard Student t test with a two-tailed distribution, using Prism GraphPad Software, version 4.0 (GraphPad Software Inc., San Diego, CA). Additional details are described in the Supporting Information.
OTHER METHODS
For identification of PDIA3 as a DKC1 regulator using a biotin-streptavidin system, liquid chromatographic–tandem mass spectrometric (LC-MS/MS) analysis, determination of gene expression levels by quantitative polymerase chain reaction, immunofluorescence assay, purification of recombinant proteins in Escherichia coli, cell viability, caspase 3/7 activity, invasion assay, protein carbonyl assay, nuclear and cytoplasmic fractionation, immunohistochemistry, disulfide isomerase activity, xenograft assay, ChIP and re-ChIP, cell culture, short interfering RNA (siRNA)–mediated gene silencing, ectopic expression of PDIA3 or DKC1 plasmids, and measurement of promoter activity, please see details in the Supporting Information.
Results
PDIA3 BINDS DKC1 REGULATORY DNA
In our previous study, we reported H2O2-mediated DKC1 up-regulation.18 Thus, in this study, to identify a regulator of the DKC1 promoter, we treated the biotin-labeled DKC1 regulatory region with nuclear extract derived from HCC cells after mock treatment and treatment with H2O2 (Fig. 1A). Comparing the pull-down proteins in mock and H2O2 treatments, we found that H2O2 treatment abolished PDIA3 binding to DKC1 (Fig. 1B,C; Supporting Fig. S1). These results suggested a role of PDIA3 in decreasing DKC1 transcriptional expression and the involvement of H2O2 in blockade of PDIA3 function in DKC1 expression repression.
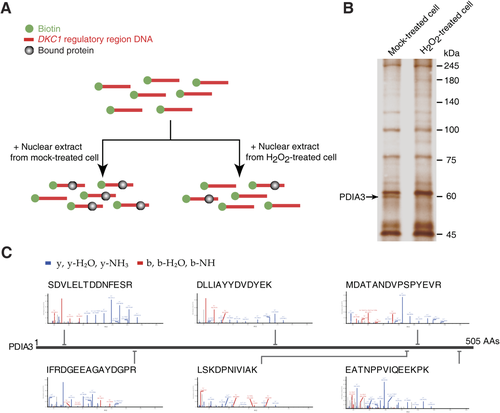
Identification of PDIA3 as a binding protein of DKC1 regulatory DNA. (A) Strategy for the identification of a transcriptional modulator of the DKC1 regulatory DNA through the biotin-streptavidin system. (B) Silver-stained gel showing the decreased binding of PDIA3 to DKC1 gene in H2O2-treated Huh7 HCC cell nuclear lysates. (C) MS peptide sequencing revealing human PDIA3 as a binding factor of DKC1 regulatory DNA. Abbreviation: AAs, amino acids.
PDIA3 ACTS AS A DKC1 TRANSCRIPTIONAL REPRESSOR IN HCC CELLS
Promoter activity of the DKC1 containing the regulatory region used in the pull-down assay for identifying transcriptional repressors was approximately 10-fold lower compared to that of the gene without the regulatory region (Fig. 2A,B). We confirmed that the decreased promoter activity was dependent on the transcriptional activity of DKC1 (Fig. 2B, middle). H2O2 increased the promoter activity of the DKC1 possessing the regulatory region used for identifying transcriptional repressors of DKC1 (Fig. 2B). Thus, we found that the locus, which included the first intron (between +118 and +744 bp; Fig. 2A), was important in regulating DKC1 transcription activity. H2O2 treatment increased DKC1 expression at both the mRNA and protein levels (Fig. 2C), while there were no changes to PDIA3 expression at either the mRNA or protein level (Fig. 2C,D). We showed that H2O2 treatment decreased the binding of PDIA3 to the DKC1 promoter region containing the first intron (Fig. 2D) and that the treatment inhibited nucleus-localized PDIA3 levels (Fig. 2E,F).
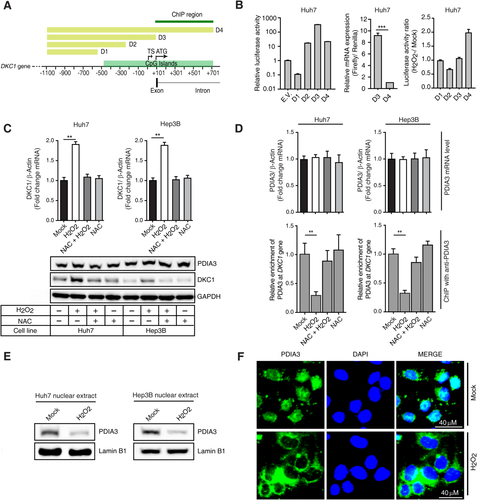
OXIDATIVE MODIFICATIONS TO PDIA3 ARE IMPORTANT FOR CONTROLLING DKC1 EXPRESSION
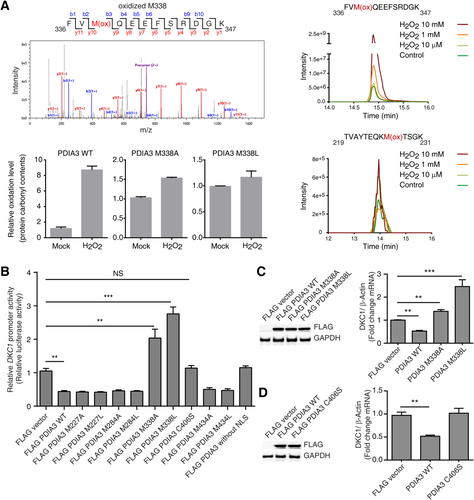
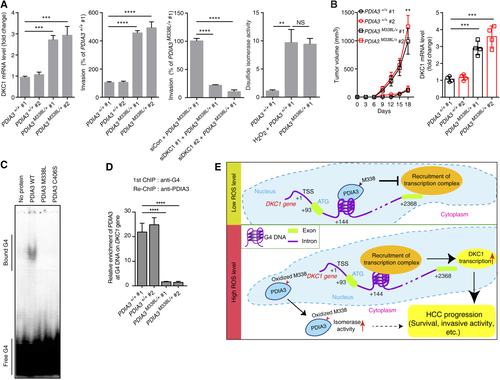
LC-MS/MS analysis of recombinant PDIA3 disclosed that residue M338 in PDIA3 was sensitively oxidized by H2O2 and that the oxidation level of this residue was positively correlated with the H2O2 concentration used for treatment, with impaired oxidation of PDIA3 protein observed in M338A and M338L mutants (Fig. 3A). However, other cysteine and methionine residues in PDIA3 were not sensitively oxidized by H2O2 compared to M338 (Fig. 3A). Although PDIA3 overexpression decreased DKC1 promoter activity and the expression level of DKC1 mRNA, the overexpression of mutant PDIA3 (M338A or M338L) increased DKC1 promoter activity and DKC1 mRNA levels (Fig. 3B,C; Supporting Fig. S21). The overexpression of the PDIA3 mutant without the nuclear localization signal (NLS) motif and the PDIA3 C406S mutant, which has an altered cysteine that is known to be critical for the DNA-binding activity of PDIA3, did not decrease DKC1 promoter activity and mRNA levels (Fig. 3B,D). Notably, in siRNA-mediated endogenous PDIA3-silenced Huh7 cells, overexpression of the PDIA3 mutant without the NLS or PDIA3 C406S increased DKC1 promoter activity (Supporting Fig. S2). Together, these results suggest that the M338L and M338A mutations increase DKC1 transcription through loss of their DKC1 gene-binding ability and inhibition of PDIA3 wild-type (WT) binding to the DKC1 gene.
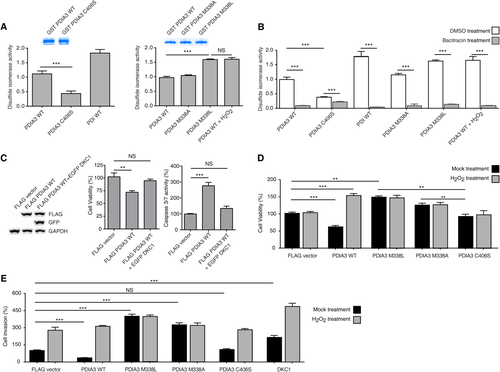
PDIA3 MUTATION M338L INCREASES DISULFIDE ISOMERASE ACTIVITY, CELL VIABILITY, AND CELL INVASION ABILITY
DKC1 mRNA levels were similar in HCC cells possessing oxidized PDIA3 induced by H2O2 and in HCC cells with PDIA3 M338L overexpression (Figs. 2C and 3C). Thus, we tested whether cytoplasmic functions were similar in PDIA3 WT and in PDIA3 M338L. Assaying the disulfide isomerase activity revealed that the activity of M338L mutant PDIA3 was similar to that of H2O2-treated PDIA3 WT, which was higher compared to that of WT (Fig. 4A). The reductase activity of C406S was lower (39% of WT activity) than that of PDIA3 WT (Fig. 4A). Bacitracin, an inhibitor of disulfide isomerase activity, induced a strong decrease in WT and M338L PDIA3 activities (90% reduction) but a moderate reduction in that of C406S (40% decrease) (Fig. 4B). Thus, disulfide isomerase activity was conserved in PDIA3 M338L mutant but reduced in C406S mutant. In addition, C406S mutation reduced the sensitivity to bacitracin, whereas M338L mutant was responsible for increasing the activity of PDIA3 in the cytoplasm. DKC1 overexpression rescued PDIA3-induced cell death because the increased caspase 3/7 activity by PDIA3 overexpression was reversed by DKC1 overexpression (Fig. 4C). These data indicate that PDIA3 activates apoptosis signaling through decreasing DKC1 expression level and sheds light on the role of PDIA3 as a DKC1 transcriptional repressor influencing tumor cell survival. Furthermore, viability was increased both in HCC cells overexpressing the M338L mutant and in H2O2-treated HCC cells overexpressing the WT (Fig. 4D). The survival rate of HCC cells overexpressing the M338L mutant was high irrespective of H2O2 exposure (Fig. 4D), suggesting that M338 acted as an essential oxidation site in PDIA3. HCC cells overexpressing WT PDIA3 also showed decreased cell invasion ability compared to HCC cells with ectopic expression of FLAG empty vector (with or without H2O2 treatment) or PDIA3 WT overexpression with H2O2 treatment (Fig. 4E). Interestingly, PDIA3 M338L overexpression increased cell invasion ability compared to that of HCC cells with DKC1 overexpression (Fig. 4E).
PDIA3 INTERACTS WITH G4 DNA STRUCTURE IN THE DKC1 REGULATORY REGION
The thioredoxin domain and glutamine–glutamate–aspartate–leucine motif are conserved in human and mouse PDIA3 (Fig. 5A). Yeast also has orthologs of PDIA3 (Fig. 5A). There were no deletion mutants showing increased yeast DKC1 mRNA levels among yeast strains harboring deletion of the PDIA3 ortholog (Fig. 5B). In addition, PDIA3 overexpression in mouse cells did not decrease DKC1 mRNA levels, indicating a possible involvement of the cis element on the DKC1 regulatory region during PDIA3-mediated DKC1 repression (Fig. 5B). However, human cell lines showed a decrease in DKC1 mRNA levels after PDIA3 overexpression (Fig. 3C). In addition, siRNA-mediated PDIA3 knockdown increased DKC1 mRNA levels (Fig. 5B). Thus, possibly, there is a particular mechanism of PDIA3 involved in regulating DKC1 transcription.
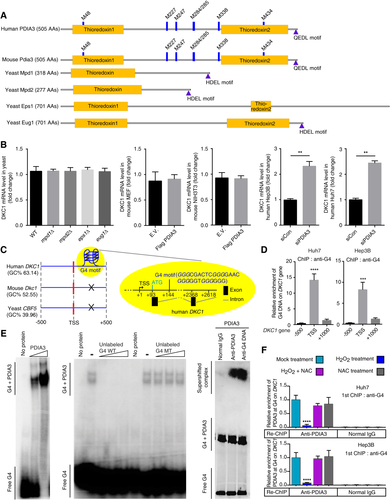
We hypothesized that DKC1 contains the elements required for modulating its transcription by PDIA3. The G4 DNA motif is located near the transcription start site of human DKC1 (Fig. 5C). We analyzed yeast, mouse, and human DKC1 using QuadBase and QGRS Mapper, and only human DKC1 had the G4 DNA element in its first intron (Fig. 5C). The G4 DNA structure contains G4-rich DNA, which is involved in transcriptional regulation.19 Using ChIP with anti-G4 antibody, we found the G4 structure near the DKC1 transcription start site in human HCC cell lines (Fig. 5D). Gel retardation assays revealed the complex formed between DNA oligomer containing DKC1 G4 motif and PDIA3 protein (Fig. 5E). ChIP using anti-G4 antibody followed by re-ChIP using anti-PDIA3 antibody revealed that H2O2 induced a sharp reduction in PDIA3 binding to the DKC1 containing G4 DNA (Fig. 5F). PDIA3 interacted with the DKC1 regulatory DNA domain by recognizing G4 DNA, and this resulted in down-regulation of DKC1 transcription.
PDIA3 MUTANT M338L INDUCES DKC1-MEDIATED TUMOR GROWTH AND INCREASES INVASION AND DISULFIDE ISOMERASE ACTIVITIES IN HCC CELLS
We tested the importance of the M338 residue in the ability of PDIA3 to modulate DKC1 expression by analyzing knock-in HCC cells harboring a heterozygous M338L mutation in PDIA3 (Supporting Fig. S3 and Tables S1-S3). Heterozygous PDIA3 M338L knock-in HCC cells (PDIA3M338L/+) were established using a CRISPR/CRISPR-associated (Cas) system, and the knock-in HCC cells showed an increase in DKC1 expression (Fig. 6A, left), invasion capacity (Fig. 6A, middle), and disulfide isomerase activity (Fig. 6A, right) of PDIA3 compared to HCC cells possessing homozygous PDIA3 WT. The heterozygous knock-in M338L mutation promoted tumor growth of immune-deficient mice, and DKC1 mRNA levels in the tumors were higher after injection of cells with the M338L mutant than after injection with PDIA3 WT (Fig. 6B). Moreover, the PDIA3 protein containing the M338L mutation showed marked reduction in G4 DNA binding (Fig. 6C,D). Thus, we concluded that PDIA3 WT recognizes the G4 DNA structure and blocks recruitment of transcriptional activators or activator-associated factors, resulting in transcriptional repression of DKC1 (Fig. 6E). The PDIA3 C406S mutant lacked the ability to bind G4 DNA (Fig. 6C), but cell viability as well as cell invasion ability were not increased by C406S overexpression. However, overexpression of PDIA3 M338L and M338A increased cell viability and invasion ability (Fig. 4D,E). The C406S mutation impaired disulfide isomerase activity, whereas PDIA3 M338L had elevated disulfide isomerase activity (Fig. 4A). These results suggest the involvement of disulfide isomerase activity in promoting HCC tumor cell progression (Fig. 6E). Moreover, our data also indicate that haplo-deficiency of functional PDIA3 is sufficient for DKC1-mediated tumor growth.
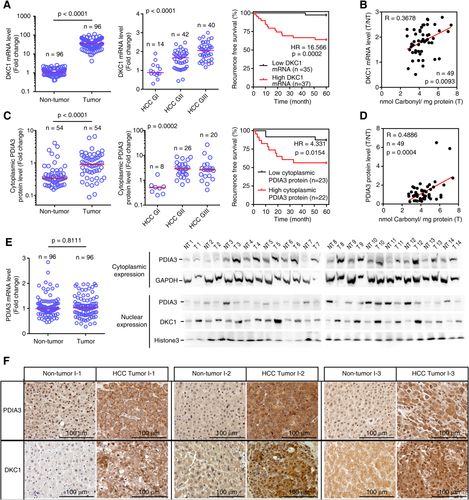
HIGH LEVEL OF CYTOPLASMIC PDIA3 EXPRESSION AND NUCLEAR DKC1 EXPRESSION CORRELATES WITH HCC PROGRESSION
DKC1 mRNA levels were higher in HCC tissues compared to those in nontumor tissue (Fig. 7A, left) and further elevated with an increase in the HCC grade (Fig. 7A, middle). Patients with HCC tumors possessing high DKC1 mRNA levels showed high HCC recurrence compared to HCC patients possessing low DKC1 mRNA levels (Fig. 7A, right), and there was a positive correlation between ROS levels and DKC1 mRNA abundance in HCC tissues derived from these patients (Fig. 7B). Cytoplasmic PDIA3 protein levels were higher in tumors compared to those in nontumors (Fig. 7C, left) and elevated with an increase in HCC grading (Fig. 7C, middle). HCC patients possessing high cytoplasmic PDIA3 levels showed high HCC recurrence compared to those possessing low cytoplasmic PDIA3 levels (Fig. 7C, right). Furthermore, cytoplasmic PDIA3 levels positively correlated with ROS levels in these patients (Fig. 7D). There was no difference in PDIA3 mRNA levels between HCC tumors and nontumors (Fig. 7E, left). However, we observed that PDIA3 protein levels were high in the cytoplasm and low in the nuclei of HCC tumors (Fig. 7E, right). The pattern of DKC1 protein expression was the opposite of that of PDIA3 protein expression in the nucleus (Fig. 7E, right). Moreover, linear regression analysis of liver tissues exhibited a significant positive correlation between nuclear DKC1 and cytoplasmic PDIA3 protein levels (Supporting Fig. S4). Immunohistochemistry revealed that HCC tumors displayed an increase in cytoplasmic PDIA3 and in nuclear DKC1 protein levels, while there was a decline in nuclear PDIA3 protein levels (Fig. 7F). Thus, the increased PDIA3 protein level was dependent on cytoplasmic PDIA3 expression, and low expression of nuclear PDIA3 induced high DKC1 expression.
Discussion
Proteins can be oxidatively modified by ROS, and cysteine and methionine moieties are particularly vulnerable to oxidation.20 Oxidative modification can change protein stability by increasing or decreasing proteolysis activity in the proteasome-dependent degradation process.21, 22 It has been suggested that oxidative modification affects protein stability differently under certain environments.20, 23-26 In addition, ROS can modulate several signal transduction pathways.20 For example, oxidation of the serine/threonine kinase protein kinase C by H2O2 stimulates prolonged kinase activity and promotes tumor growth.20, 24 H2O2 also induces oxidative modification of the phosphatase and tensin homolog deleted on chromosome 10, which interferes with its role as a phosphoinositide 3-kinase antagonist.23 Thus, oxidative modification of proteins is important for regulation of intracellular events.
Reports have shown that PDIA3 levels were high in the sera of patients with HCC as well as in HCC tissues compared to those in healthy liver or nontumors.27, 28 Interestingly, we found that PDIA3 is also oxidatively modified in HCC cells under high ROS levels, resulting in its redistribution and changes in its function. Among the oxidative modifications of PDIA3, we found that modification of M338 is important for changes in PDIA3 functions. The 1000 Genomes Project (http://www.1000genomes.org) also confirmed the possibility that the M338 residue is critical for PDIA3 function. The PDIA3 M338L mutation, where M338 was mutated to leucine, was identified as rs756473375 variant based on the 1000 Genomes Project. This mutation has been predicted to be “probably damaging” with a score of 0.995 (score range 0.000-1.000) by the PolyPhen program (http://genetics.bwh.harvard.edu/pph2/index.shtml). Whether M338L is associated with a disease and has clinical significance is currently unknown. In our data, the PDIA3 M338L mutation resulted in loss of G4 DNA binding activity. Eventually, PDIA3 modification contributed to HCC malignancy through the regulation of DKC1 transcription levels. This M338L mutation also increased PDIA3 isomerase activity. Our data show that the decrease in cell viability and invasive activity of HCC cells through PDIA3 overexpression was mostly recovered by DKC1 overexpression. We also show that in siRNA-mediated, PDIA3-silenced Huh7 cells, the DKC1 promoter activity of overexpression of PDIA3 M338L or M338A mutant was similar to that of overexpression of PDIA3 mutant without the NLS motif or that of the C406S mutant, which lacks DNA-binding ability; moreover, DKC1 knockdown resulted in sharp reduction of invasion ability in PDIA3 M338L knock-in HCC cells. These results indicate that nuclear PDIA3 inhibited DKC1 transcription and tumor progression.
DKC1 is a component of the telomerase holoenzyme, and this protein is frequently up-regulated in many human cancers.1, 4 Thus, tight regulation of DKC1 expression is required for preventing tumorigenesis. Interestingly, our sequence analysis of the DKC1 regulatory region showed that the G4 DNA region was located in the first intron of DKC1 and that only human DKC1 possesses the regulatory region; this indicates that the addition of an extra regulatory region in human DKC1 occurred during evolution. G4 DNA can be created and sustained in 5′ untranslated regions (UTRs), first introns, and 3′ UTRs in a stable state during replication, transcription, and recombination in living cells.29 The G4 sequence is reportedly located within the 5′-most 100 bp of the first intron.29 The G4 sequence can be involved in regulation of gene expression; for example, G4 DNA causes RNA polymerase II pausing, and G4-stabilizing ligands, such as pyridostatin, inhibit gene transcription.30, 31 Our data suggest that the G4 DNA region in DKC1 was affected by ROS and was important for transcriptional regulation. In this regulation, we found that PDIA3 binding to G4 DNA in DKC1 was critical and that DNA binding activity of PDIA3 was affected by ROS-mediated oxidative modification at a specific residue (M338). Thus, in this report, we elucidated the mechanism underlying repression of DKC1 transcription by PDIA3 through binding to G4 DNA.
In this report, we demonstrated oxidative modification of M338 of PDIA3 and revealed that the M338 site primarily contributes to PDIA3 action in DKC1 transcriptional regulation. In addition, we observed that oxidatively modified PDIA3 accumulated in the cytoplasm of HCC cells, resulting in elevated DKC1 expression. Malignant HCC tissues also showed increased cytoplasmic PDIA3 abundance and increased nuclear DKC1 expression. Thus, we conclude that PDIA3 with oxidatively modified M338 increases DKC1 mRNA levels and tumor cell survival; consequently, PDIA3 drives progression of malignant tumors in the liver.
Acknowledgment
The biospecimens were provided by the Korea University Guro Hospital of National Biobank and the Inje Biobank of Inje University Busan Paik Hospital, a member of the Korea Biobank Network. We are deeply thankful to Dr. Eun Sun Jung, a pathologist at Seoul St. Mary's Hospital, for valuable advice and immunohistochemical analysis, and Dr. Ji Yoon Lee, a senior researcher at the National Instrumentation Center for Environmental Management at Seoul National University for LC-MS/MS analysis for PDIA3 identification.
REFERENCES
Author names in bold designate shared co-first authorship.




