Multiplatform Genomic Roadmap of Hepatocellular Carcinoma: A Matter of Molecular Heterogeneity
Supported by grants from the National Research Foundation of Korea (NRF) funded by the Korean government (NRF-2017R1E1A1A0 1074733, NRF-2017M3A9B6061509) and by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea (HI16C1074).
Over the past decade, large-scale international initiatives in genomics, such as The Cancer Genome Atlas Research Network (TCGA) and the International Cancer Genome Consortium (ICGC), have generated open-access multiomics profiles revealing the genomic landscapes of cancers. For hepatocellular carcinoma (HCC), TCGA researchers published a first landmark multiomics study using the six different platforms of DNA somatic mutations and copy number aberrations in 363 samples, and DNA methylation, proteomic, mRNA, and microRNA profiles in 196 samples.1 The data provided knowledge resources that facilitate our understanding of cancer biology and have led to the identification of potential new diagnostic or therapeutic targets for precision medicine.
In-depth genomic analyses of TCGA data could validate recurrent mutations in a handful of driver genes and genomic aberrations that have been known to occur during HCC progression. In particular, mutations were detected in the tumor-suppressor genes TP53 (tumor protein p53; 31% of samples), AXIN1 (8%), and RB1 (4%), the oncogene CTNNB1 (catenin beta 1; 27%), and the chromatin remodeling genes ARID1A (7%), ARID2 (5%), and BAP1 (BRCA1-associated protein 1; 5%). The prognostic significance of signatures related to isocitrate dehydrogenase 1 and 2 (IDH1/2) and TP53 mutations has also been investigated.
The most important feature of the TCGA study is its simultaneous generation of multiple types of omics profiles, including genomics, epigenomics, transcriptomics, and proteomics, in the same samples. By performing integrative clustering analysis of multiomics profiles, researchers have defined three molecularly distinct subtypes, iClust1, 2, and 3, that may differ biologically and therefore contain distinctive drug targets. This molecular taxonomy provides biological insights into the complex, multilayered interactions among DNA, RNA, and proteins in cancer cells, which may reflect the mechanistic architecture underlying HCC heterogeneity. Although the classification was not related to overall survival, the subtypes overlapped substantially with previous molecular classifications based on gene expression profiling.2 In comparison with the iClust2 and iClust3 subtypes, the iClust1 subtype exhibited several poor prognostic features, including hepatic stem cell–like and cholangiocarcinoma-like, and other prognostic expression traits. The iClust1 subtype was also characterized by low frequencies of cyclin dependent kinase inhibitor 2A (CDKN2A) silencing, CTNNB1 mutation, and telomerase reverse transcriptase (TERT) promoter mutation, as well as high expression of microRNA (miR)-181a and epigenetic silencing of miR-122. Interestingly, the TCGA study found IDH1/2 mutations in iClust1, linking hypermethylation to the expression of stem cell–like and cholangiocarcinoma-like traits. IDH1/2 mutations were recurrently detected in intrahepatic cholangiocarcinoma (iCCA), implying that such mutations are a common genetic feature of HCC and iCCA. Previously, the common features between HCC and iCCA have been noticed to have functional and prognostic significance.3
In parallel to the TCGA study, two other multiomics studies of HCC were published in 2017. They also reported multiomics-based molecular subclasses in different patient cohorts, revealing different key drivers of HCC progression. Comparison of HCC and iCCA in cohorts from Thailand revealed that common features of these tumor types (e.g., PLK1 and ECT2) play key roles in cancer progression.4 In another study by our group, we determined the correlated gene features among the multiomics data, which represent potential interacting genes. Multilayered genomic and epigenomic aberrations of these genes could identify three molecular subtypes. One of the subtypes was characterized by recurrent mutation of BAP1, which is frequently mutated in iCCA, and concomitant expression of stem cell–like traits such as CA-9 and KRT19.5 These studies, along with the TCGA study, consistently suggest that the common features of HCC and iCCA play critical roles in acquisition of the aggressive HCC phenotype, linking their resemblance with bipotential stem cells that can differentiate into hepatocytes as well as cholangiocytes.
Such efforts toward molecular classification can provide deeper insights into the pathobiology of cancers. However, no consensus currently exists regarding the precise molecular targets of HCC that are most readily applicable in the clinic. In part, this is because the marker genes defining each subclass differ across studies. One possible reason for these discrepancies is that the classifications are largely based on transcriptomic features. Because the transcriptome profiles are overly tumor-centric, they may underestimate the effects of inflammatory or cirrhotic microenvironments. Nontumoral tissues may provide complex microenvironment interacting with surrounding HCCs, including immune cell activities, hypoxic and metabolic status, and/or stromal cell activities. The interaction between them may contribute to the definite alternation of tumor behaviors. In light of this concern, it is notable that TCGA performed transcriptome analysis specifically in regard to immune signals and confirmed the association between CTNNB1-activating mutations and a lack of immune infiltration, which has been addressed.6 A molecular-level understanding of the inflammatory milieu may yield more informative biomarkers for immune therapy, which, in turn, might enable prediction of HCC subtypes with higher confidence.
On the other hand, it should be taken into account that HCC patients have a huge variety of clinical backgrounds, such as etiology, race/ethnicity, and geographical distributions. Large-scale collection of TCGA samples inevitably includes heterogeneous cohorts from different backgrounds, which can affect the molecular classification. For example, the subtype iClust1 was clinically associated with young age, Asian ethnicity, female sex, and normal body weight. It has been reported that TP53 mutation frequency in HCC is highly associated with its etiology and geographical distribution: TP53 mutations were present in 20%-50% of HCCs in China, but in only 5%-20% of HCCs in Europe and other Asian countries such as Korea and Japan.7, 8 This discrepancy might be related to differences in etiological prevalence across countries. Notably in this regard, the core sample set of the TCGA data (n = 196) had a heterogeneous ethnic composition, including 117 white (59.7%), 55 Asian (28%), and 14 black (7%) subjects. Even a single racial group (Asian) consisted of patients from different countries, including Vietnam (n = 26), Korea (n = 4), and Canada (n = 25), which may affect mutation frequencies. Indeed, in comparison with patients from other countries, alcohol-related etiology was highly prevalent in Vietnamese patients (14 of 26; 53.8%), as were recurrent TP53 mutations (13 of 26; 50%). Thus, careful consideration might be required on the sample composition from various cohorts, particularly in the context of mutation analysis.
The TCGA study represents a tremendous data resource for HCC genomics and provides many intriguing clues regarding the heterogeneous pathobiology of HCC progression. Undoubtedly, these resources will help to catalyze future mechanistic studies of HCC, leading to discoveries of novel precision-medicine targets and diagnostics for patient management. For future studies, the extraction of biologically relevant information from complex, multidimensional cancer data sets remains a significant challenge for new cancer target discovery. Huge molecular heterogeneity and complexity of HCC might be achieved by combinatorial explosion of interactions among the multilayered genomic and epigenomic aberrations and their shaping by environmental influences (Fig. 1). Thus, although TCGA produced the largest data set related to the HCC genomic repertoire and associated clinical information, the sample size seems not enough to resolve the heterogeneity of this disease. The primary challenge of HCC still lies in the characterization of its heterogeneity and complexity. Expanded and selected sample collection, taking into account etiology, ethnicity, and other clinic-pathological features, might be necessary in order to clarify the HCC heterogeneity.
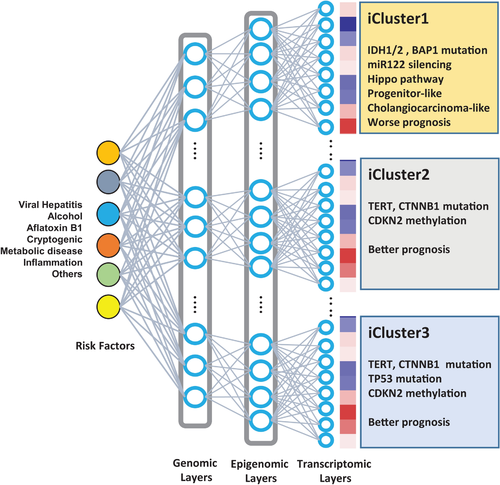
Multilayered interaction of the genomic and epigenomic aberrations in HCC. The figure illustrates the combinatorial interactions among the genomic and epigenomic aberrations that give rise to heterogeneous progression of HCC. The characteristics of the molecular subtypes are shown.
In addition, intratumoral heterogeneity in the same patients was not addressed in the TCGA study. HCCs have shown considerable variation even within a same patient; therefore, sequence analysis of a single lesion may not represent the genomic features of the patients.9 Recent advances in single-cell sequencing technology will increase the resolution of genome data, providing new insights into the pathobiology of intra- and intertumoral heterogeneity, as well as the interplay between tumors and cells in the microenvironment. Moreover, efforts to unite molecular classifications with pathological and/or radiological features might be a plausible strategy to define more-robust patient subclasses, because tumor behaviors are also strongly linked with morphological and/or pathological features. Multiomics profiling of multistep hepatocarcinogenesis will also promise to elucidate pathological derivation that promote malignant conversion of precancerous lesions.
Furthermore, to move closer to clinical application, enormous translational efforts are required. The TCGA study is mainly aimed to provide biological insights on molecular heterogeneity in a large-scale data; they lack enough information about clinical courses of the patients such as drug responses, therapeutics, or disease progression with longitudinal follow-up. Further data collection focusing on such clinical outcomes might be required.
It is time to demonstrate that precision biomarkers defining subclasses of patients will ultimately lead to better therapies in which the clinical decision-making process is influenced by the molecular heterogeneity of HCC. In the era of precision medicine, actionable targets may not necessarily be recurrent events in cancer patients. Rare variants, even with a singleton observation, may also be considered as potential targets for companion diagnostics and/or targeted therapies if they can be validated as actionable.
In conclusion, the data resources from the TCGA study will undoubtedly convey a further explosion of findings in the HCC field, which will accelerate the realization of precision medicine.
Potential conflict of interest
Nothing to report.




