H19 promotes cholestatic liver fibrosis by preventing ZEB1-mediated inhibition of epithelial cell adhesion molecule
Potential conflict of interest: Nothing to report.
Li Wang was supported by the National Institutes of Health (grants R01DK104656, R01DK080440, R01ES025909, R21AA022482, and R21AA024935); VA Merit Award 1I01BX002634; the Yale Liver Center (grant P30 DK34989) and the National Natural Scientific Foundation of China (grant 81572443). Olivier Barbier was supported by the Canadian Institute of Health Research (grant 119331), the Canadian Foundation for Innovation (grant 25712), the Canadian Liver Foundation, and the Natural Sciences and Engineering Research Council of Canada.
Abstract
Based on our recent finding that disruption of bile acid (BA) homeostasis in mice results in the induction of hepatic long noncoding RNA H19 expression, we sought to elucidate the role of H19 in cholestatic liver fibrosis. Hepatic overexpression of H19RNA augmented bile duct ligation (BDL)-induced liver fibrosis, which was accompanied by the elevation of serum alanine aminotransferase, aspartate aminotransferase, bilirubin, and BA levels. Multiple genes related to liver fibrosis, inflammation, and biliary hyperplasia were increased in H19-BDL versus null-BDL mice, whereas genes in BA synthesis were decreased. Livers and spleens of H19-BDL mice showed significant enrichment of CD3+γδ+, interleukin-4, and interleukin-17 producing CD4+ and CD8+ immune cell populations. H19 down-regulated hepatic zinc finger E-box-binding homeobox 1 (ZEB1) but up-regulated epithelial cell adhesion molecule (EpCAM) and SRY (sex determining region Y)-box 9 expression. Mechanistically, ZEB1 repressed EpCAM promoter activity and gene transcription. H19RNA impeded ZEB1's inhibitory action by interacting with ZEB1 protein to prevent its binding to the EpCAM promoter. Hepatic overexpression of ZEB1 or knockdown of EpCAM diminished H19-induced fibrosis; the latter was also prevented in H19−/− mice. H19RNA was markedly induced by bile acids in mouse small cholangiocytes and to a lesser extent in mouse large cholangiocytes. The up-regulation of H19RNA and EpCAM correlated positively with the down-regulation of ZEB1 in primary sclerosing cholangitis and primary biliary cirrhosis liver specimens. Conclusion: The activation of hepatic H19RNA promoted cholestatic liver fibrosis in mice through the ZEB1/EpCAM signaling pathway. (Hepatology 2017;66:1183-1196).
Abbreviations
-
- AAV8
-
- adeno-associated viral vector serotype 8
-
- ALT
-
- alanine aminotransferase
-
- AST
-
- aspartate aminotransferase
-
- α-SMA
-
- alpha smooth muscle actin
-
- BA
-
- bile acid
-
- BDL
-
- bile duct ligation
-
- Ck7
-
- cytokeratin 7
-
- Ck19
-
- cytokeratin 19
-
- Col1a1
-
- collagen type I alpha 1
-
- Col1a2
-
- collagen type I alpha 2
-
- EpCAM
-
- epithelial cell adhesion molecule
-
- EMT
-
- epithelial-to-mesenchymal transition
-
- Fgfr4
-
- fibroblast growth factor receptor 4
-
- Fxr
-
- farnesoid X receptor
-
- H&E
-
- hematoxylin and eosin
-
- lncRNA
-
- long noncoding RNA
-
- MLC
-
- mouse large cholangiocyte
-
- MSC
-
- mouse small cholangiocyte
-
- mRNA
-
- messenger RNA
-
- PBC
-
- primary biliary cirrhosis
-
- PSC
-
- primary sclerosing cholangitis
-
- qPCR
-
- quantitative polymerase chain reaction
-
- SEM
-
- standard error of the mean
-
- SHP
-
- small heterodimer partner
-
- SOX9
-
- SRY (sex determining region Y)-box 9
-
- ZEB1
-
- zinc finger E-box-binding homeobox 1
Bile acids (BAs) can facilitate intestinal absorption and transportation of lipids and other nutrients. Impairment of bile formation or bile flow to the gallbladder and duodenum results in cholestatic liver disease.1 Cholestatic liver fibrosis develops due to the activation of matrix-producing hepatic stellate cells, which is signified by excessive extracellular matrix deposition and hepatic stellate cell differentiation into myofibroblast-like cells.2 Surgical ligation of the common bile duct (BDL) is a commonly used model to induce obstructive cholestatic injury in rodents, which disrupts bile flow and results in the accumulation of BAs in the liver.3
Long noncoding RNAs (lncRNAs) represent a group of transcripts that are above 200 nucleotides in length but without protein coding potential. As one of the first identified lncRNAs, the H19 gene is paternally imprinted, therefore it is only transcribed from the maternally inherited allele.4 H19 has been recognized with a broad spectrum of functions in multiple physiological and pathological processes.4 However, its regulatory function in the liver remains largely unknown. Our recent study showed that hepatic H19RNA was drastically induced in Bcl2-induced cholestatic liver injury as well as in human liver specimens from patients with fibrosis and cirrhosis,5 indicating a clinically relevant regulatory role of H19 in chronic liver disease.
Zinc finger E-box binding homeobox 1 (ZEB1) is a transcriptional repressor that has been implicated in epithelial-to-mesenchymal transition (EMT) during tissue fibrosis.6 ZEB1 inhibits E-cadherin and plays a major role in triggering EMT during organ fibrosis and cancer cell metastasis.7 Its expression is controlled by the microRNA-200 family6 and is modulated by nuclear receptors small heterodimer partner (SHP) and liver receptor homolog-1.8 Interestingly, the H19 gene transcription is inhibited by SHP,5 a critical negative regulator of BA synthesis and cholestasis,9, 10 and H19 is also involved in the regulation of EMT.11 These studies suggest a potential cross-talk between H19 and ZEB1 in the control of cholestatic liver fibrosis.
The epithelial cell adhesion molecule (EpCAM, also known as CD326) is a homophilic Ca2+-independent cell–cell adhesion molecule that is expressed in many human epithelial tissues. It is a transmembrane glycoprotein that is frequently overexpressed in embryonic stem cells, tissue progenitors, carcinomas, and cancer-initiating cells.12 EpCAM is associated with enhanced cell proliferation, and it transmits signals into nucleus through intramembrane proteolysis to release and promote nucleus translocation of its intracellular domain EpICD.13, 14 Despite intensive studies focusing on the role of EpCAM in various cancers, little is known about its regulation by lncRNA. Interestingly, in a rodent model of HCC induced by diethylnitrosamine and carbon tetrachloride, both H19RNA and EpCAM messenger RNA (mRNA) were increased in liver tumors.15 This raises the possibility for a common pathway regulated by H19 and EpCAM in liver disease.
The present study was undertaken to understand the function of H19 in cholestasis. Our study identified a H19/ZEB1/EpCAM cascade as a new regulatory module to control the development of cholestatic liver injury and fibrosis.
Materials and Methods
ANIMALS AND HUMAN LIVER SPECIMENS
C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). H19−/− mice were kindly provided by Linheng Li.16 Because H19 is a paternal imprinted gene, maternal H19-deleted mice were used for experiments and paternal H19-deleted mice were used as negative controls. Mice were fed a standard rodent chow diet (Harlan No. 2018), had free access to water, and were maintained in a 12-hour light/dark cycle in a temperature-controlled (23°C) and pathogen-free facility. In vivo experiments were performed on male mice at the age of 6 weeks unless stated otherwise (n = 5 mice/group). For in vivo viral transduction,17 mice were injected via the tail vein with purified adeno-associated viral vector serotype 8 (AAV8) containing a liver-specific thyroxine-binding globulin promoter driving H19 gene expression at 5 × 1010 virus particles per mouse. Sham and BDL surgeries were performed for 1 week post-AAV8-null or AAV8-H19 transduction for 1 month, using our established method.3 For rescue experiments, mice with H19 overexpression for 1 month were injected via the tail vein with mouse pShuttle-ZEB1 overexpression plasmid (GeneCopoeia, Rockville, MD), Epcam short hairpin RNA plasmid (ShEpcam Sigma-Aldrich, St. Louis, MO), or control plasmid at 50 μg per mouse using TurboFect in vivo Transfection Reagent (R0541; Thermo Scientific, Waltham, MA) as described previously.18 All mice were sacrificed after overnight fasting unless otherwise indicated. Basic procedures to analyze animal metabolic phenotypes and serum parameters have been described previously.19 The hepatic bile acid (BA) pool, intestine bile pool, fecal BA excretion and serum BA concentration were measured with a Colorimetric Total Bile Acid Assay Kit (CELL BIOLABS, Inc) as described previously.20 Hematoxylin and eosin (H&E) and Masson trichrome staining were performed, and eight fields of each slide were randomly placed under a microscope and quantified by ImageJ software as described previously.5 Protocols for animal use were approved by the Institutional Animal Care and Use Committee at the University of Connecticut.
The coded normal human liver specimens, primary sclerosing cholangitis (PSC), and primary biliary cirrhosis (PBC) were obtained through the Liver Tissue Cell Distribution System (Minneapolis, MN), which was funded by National Institutes of Health Contract HSN276201200017C. The patient information of PBCs and PSCs are shown in Supporting Table S1. Because we did not ascertain individual identities associated with the samples, the Institutional Review Board for Human Research at the University of Connecticut determined that the research did not involve human subjects.
CELL LINES, PLASMIDS AND ANTIBODIES
The human hepatoma cell lines HepG2,5 Huh7,21 Hep3B22; mouse hepatoma cell line Hepa123; human embryonic kidney 293T24; mouse large cholangiocytes (MLCs); mouse small cholangiocytes (MSCs)25; human immortalized, nonmalignant cholangiocyte cell line H69; and four human cholangiocarcinoma cell lines Mz-Cha-1, CCLP-1, HuCC-T1, and SG231 with different biliary origin26 have been described previously. ZEB1 3′-untranslated region (3′-UTR) luciferase construct was obtained from Addgene (Cambridge, MA).6 ZEB1 CRISPR activator and CRISPR KO constructs were purchased from Santa Cruz Biotechnology (Dallas, TX). Epcam-Luc construct was kindly provided by Xinwei Wang (National Cancer Institute, Bethesda, MD).27 The mutant plasmid was constructed in our laboratory using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Ipswich, MA).28 The antibodies were purchased from Sigma-Aldrich (ZEB1, alpha smooth muscle actin [α-SMA]), Abcam (Cambridge, United Kingdom; EpCAM), Cell Signaling Technology (Danvers, MA; p-Akt, Akt, p-ERK, ERK, SMAD2/3, β-Actin), and Santa Cruz Biotechnology (α-Tubulin). Transcription factor and promoter interaction was predicted by Jaspar (http://jaspar.genereg.net/). RNA pull-down was performed as described previously.20
OTHER STANDARD METHODS
RNA isolation, quantitative polymerase chain reaction (qPCR), cell culture, in vitro virus transduction and plasmid transfection, western blot analysis, and confocal imaging have been described previously.17, 29-31 Chromatin immunoprecipitation-qPCR, transient transfection, and luciferase reporter assay have been described previously.3, 20, 32, 33 For western blot analysis, equal amounts of proteins from five mouse livers in each group (n = 5/group) were pooled, and single or duplicate loading was used. The primers used are shown in Supporting Table S2. BA composition was analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS), as described previously.20 Detailed methods can be found in the Supporting Information.
STATISTICAL ANALYSIS
Data are shown as the mean ± standard error of the mean (SEM) and are representative of at least three independent experiments. Statistical analysis was performed using a Student t test between two groups and one-way analysis of variance among multiple groups; P < 0.05 was considered statistically significant.
Results
H19 OVEREXPRESSION FACILITATED THE DEVELOPMENT OF OBSTRUCTIVE CHOLESTATIC LIVER FIBROSIS
To elucidate the in vivo function of H19 in BDL induced-cholestatic liver fibrosis, we generated H19 overexpressed mouse models via tail vein injection of AAV8-H19 or AAV8-null (scramble control) virus driven by a specific promoter thyroxine-binding globulin for hepatic-directed delivery (Supporting Fig. S1); an established approach with long-lasting effect without toxicity.
It was well characterized that WT mice subjected to BDL for 14 days developed severe cholestatic liver fibrosis.5 We conducted sham or BDL surgery on null and H19 mice for 7 days to determine whether H19 worsened BDL-induced liver injury. H&E staining revealed liver necrosis in null-BDL mice, which was exacerbated in H19-BDL mice (Fig. 1A, left). In addition, severe liver fibrosis and bile duct proliferation were observed in H19-BDL versus null-BDL mice, as determined by Masson's trichrome and sirius red staining (Fig. 1A, right, and Supporting Fig. S2) and quantified using ImageJ software (Fig. 1B). H19 mRNA was induced by BDL and the expression of fibrogenic genes collagen type I alpha 1 (Col1a1) and collagen type I alpha 2 (Col1a2) was noticeably increased by BDL versus sham in null mice and was further elevated in H19 BDL mice (Fig. 1C).
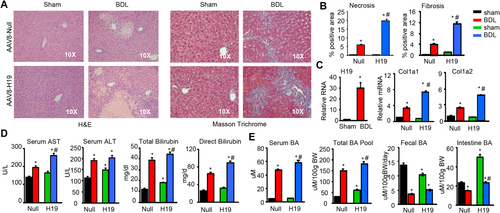
Serum aspartate aminotransferase (AST), alanine aminotransferase (ALT), total bilirubin, and direct bilirubin levels were significantly higher in null BDL versus null sham mice, which remained at high levels or were further increased, but to various extents, in H19 BDL versus null BDL mice (Fig. 1D). Serum BA and BA pool size were also largely increased in null BDL versus null sham mice and were further elevated in H19 BDL mice (Fig. 1E). In contrast, fecal BA excretion and intestine BA levels were lower in null BDL versus null sham mice. Interestingly, H19 sham mice had decreased fecal BA secretion but showed much higher levels of intestine BA; both of which were decreased in H19 BDL mice. Overall, the results suggest that overexpression of H19 exacerbated BDL-induced obstructive cholestatic liver fibrosis by disrupting BA homeostasis.
H19 INDUCED EpCAM BUT INHIBITED ZEB1 EXPRESSION IN BDL MICE
To elucidate the molecular basis by which H19 disrupted BA homeostasis, we analyzed major regulators in the BA synthesis pathways. The mRNA expression of farnesoid X receptor (Fxr) and fibroblast growth factor receptor 4 (Fgfr4) was induced by BDL in null mice versus sham control, however, such induction was blunted in H19 BDL compared with null BDL mice (Fig. 2A). Cyp7a1, a rate-limiting enzyme in BA synthesis, showed an expression pattern similar to that of Fxr and Fgfr4. Interestingly, two other BA synthetic enzymes, Cyp8b1 and Cyp7b1, exhibited expression patterns distinct from that of Cyp7a1; they were induced in H19 sham versus null sham but decreased in both null BDL and H19 BDL mice. Although Shp is an Fxr target gene, its expression was down-regulated in null BDL versus null Sham mice. Several genes in multiple signaling pathways also displayed noticeable changes. Specifically, Epcam, FBJ osteosarcoma oncogene (Fos), chemokine (C-C motif) receptor 2 (Ccr2), cytokeratin 19 (Ck19), and cytokeratin 7 (Ck7) were all activated in null BDL versus null sham and further activated in H19 BDL relative to null BDL (Fig. 2B).
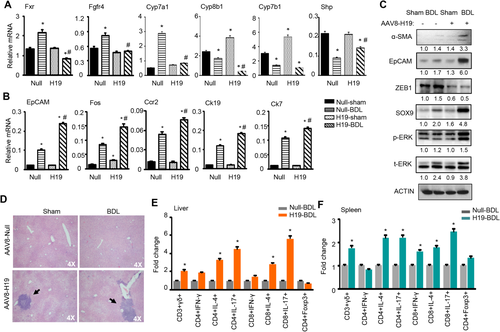
Western blot revealed a modest elevation of α-SMA protein in null BDL versus null sham, but a greater induction in H19 BDL (Fig. 2C); the latter was consistent with a more severe fibrosis observed in these mice. The expression pattern of EpCAM and SRY (sex determining region Y)-box 9 (SOX9) proteins was similar to α-SMA protein, suggesting their positive association with fibrosis in H19 BDL. On the contrary, ZEB1 protein was diminished in H19 sham versus null sham, which remained at constant low levels in H19 BDL versus null BDL mice. p-ERK protein was also elevated in H19 BDL mice. Interestingly, the expression of AKT and SMAD2/3 proteins did not show significant changes in H19 BDL versus null BDL mice (Supporting Fig. S3). Considering the transcriptional function of ZEB1, we reasoned that the decreased ZEB1 may be associated with the up-regulation of EpCAM in H19 BDL mice.
Another important observation was significant immuno-cell infiltration in H19-BDL mice (Fig. 2D). FACS analysis of subsets of immune components revealed that CD3+γδ+ cell population in livers and spleens from H19-BDL mice was markedly enriched compared with null-BDL mice (Fig. 2E,F and Supporting Fig. S4). Interleukin-4– and interleukin-17–producing CD4+ and CD8+ cell populations in livers and spleens from H19-BDL mice were also up-regulated versus null-BDL mice. There was no difference in interferon-γ–producing CD4+ and CD8+ cell populations in livers of both groups, although the interferon-γ–producing CD8+ cell population in spleens of H19-BDL mice was up-regulated. In addition, the Foxp3+CD4+Treg subpopulation showed no difference between H19-BDL and null-BDL mice. These results suggest a potentially important role of H19 in regulating hepatic immunoresponse.
H19 IMPEDED ZEB1-MEDIATED TRANSCRIPTIONAL INHIBITION OF EpCAM
To elucidate the regulation of EpCAM by ZEB1, we overexpressed ZEB1 in human HepG2 cells and observed a reduction of EpCAM protein and mRNA by ZEB1 (Fig. 3A). Because the transfection efficiency of HepG2 cells was low, we used mouse Hepa1 and human Huh7 cells for knockdown experiments. Both EpCAM protein and mRNA were markedly induced in Hepa1 and Huh7 cells transfected with shZEB1 compared with controls, and a more striking effect was observed in Huh7 cells (Fig. 3B). These results suggest that ZEB1 inhibited EpCAM expression, likely at the transcriptional level.
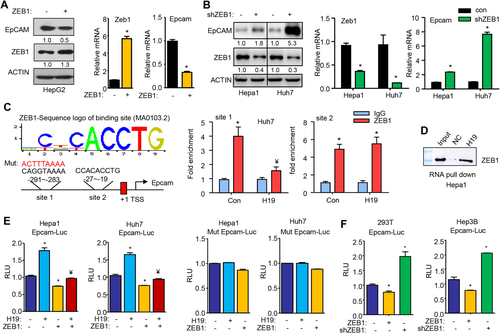
We analyzed EpCAM promoter and identified two putative ZEB1 binding sites within the proximal promoter region (Fig. 3C, left). Chromatin immunoprecipitation assays revealed a marked enrichment of ZEB1 protein to both site 1 and site 2 in the endogenous EpCAM promoter in Huh7 cells (Fig. 3C, right). Interestingly, overexpression of H19 inhibited ZEB1 binding to site 1 without affecting its binding to site 2, suggesting a site-dependent selectivity. We next conducted RNA pull-down assays combined with western blot analysis. In vitro transcribed H19RNA probe and negative control RNA probe were incubated with Hepa1 cell lysis to detect the interaction between H19RNA and the endogenous ZEB1 protein. The result showed that the H19RNA probe recruited a significant amount of ZEB1 protein than negative control, indicating an association between H19 and ZEB1 (Fig. 3D). Thus, the direct H19RNA–ZEB1 protein interaction likely resulted in dissociation of ZEB1 protein from site 1 in the EpCAM promoter.
Promoter luciferase reporter assays further demonstrated that EpCAM promoter activities in Hepa1 and Huh7 cells were activated by overexpression of H19 but reduced by overexpression of ZEB1 (Fig. 3E, left two panels). In contrast, the effect of H19 was completely blunted and the inhibition of ZEB1 was largely impaired when site 1 was mutated in the EpCAM promoter (Fig. 3E, right two panels). We subsequently examined EpCAM promoter activity in two additional cell lines 293T and Hep3B cells and observed an inhibition by ZEB1 overexpression and activation by knockdown of ZEB1 (Fig. 3F). Taken together, these results provide convincing evidence that ZEB1 served as a transcriptional repressor of EpCAM expression and that H19 activated EpCAM by dissociating ZEB1 from the EpCAM promoter.
OVEREXPRESSION OF ZEB1 OR KNOCKDOWN OF EpCAM ALLEVIATED FIBROSIS IN H19 BDL MICE
We conducted rescue experiments to determine the role of ZEB1 and EpCAM in H19-mediated cholestatic fibrosis. Mice transduced with AAV8-H19 for 1 month were injected with Zeb1, shEpcam, or control plasmid via the tail vein for 1 day followed by BDL for 7 days. H&E and Masson's trichrome staining revealed significantly attenuated fibrosis in H19-Zeb1-BDL and H19-shEpcam-BDL mice compared with H19-Con-BDL mice (Fig. 4A). The reduced liver injury was accompanied by the decreased serum ALT, AST, and bilirubin levels (Fig. 4B).
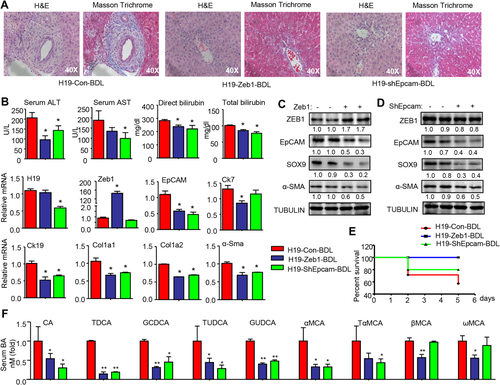
Interestingly, hepatic H19RNA was decreased in shEpcam-BDL mice, suggesting a potential feedback activation of H19 by EpCAM (Fig. 4B). As expected, Epcam mRNA was reduced in Zeb1-BDL mice. In addition, the expression of cholangiocyte markers (Ck7, Ck19) and fibrosis markers (Col1a1, Col1a2, α-Sma) was markedly decreased in Zeb1-BDL and shEpcam-BDL mice.
Western blot confirmed the decreased EpCAM proteins in Zeb1-BDL livers (Fig. 4C), providing in vivo evidence that EpCAM was the direct target of ZEB1. It was noted that ZEB1 proteins remained unchanged in shEpcam-BDL mice (Fig. 4D), suggesting that EpCAM was the downstream gene responsible for promoting cholestatic liver fibrosis. As expected, SOX9 and α-SMA proteins were decreased in both Zeb1-BDL and shEpcam-BDL mice. The survival curve showed that the mortality rates of H19-Zeb1-BDL and H19-shEpcam-BDL mice were markedly lower than that of H19-BDL mice (Fig. 4E). Analysis of serum bile acid composition revealed decreased levels of cholic acid (CA), taurodeoxycholic acid (TDCA), glycochenodeoxycholic acid (GCDCA), tauroursodeoxycholic acid (TUDCA), and glycoursodeoxycholic acid (GUDCA) in Zeb1 and shEpcam mice compared to H19 mice (Fig. 4F). In addition, CDCA-derived alpha-muricholic acid (αMCA) and its conjugate TαMCA were also decreased in Zeb1 and shEpcam mice. Beta-muricholic acid (βMCA) and its derived omega-muricholic acid (ωMCA) were only diminished in Zeb1 but not in shEpcam mice. Overall, either overexpression of ZEB1 or knockdown of EpCAM rescued liver fibrosis induced by H19 by altering BA composition.
H19-DEFICIENCY PREVENTED CHOLESTATIC LIVER FIBROSIS
We further elucidated the role of H19 in BDL-induced cholestasis by using H19−/− mice. As a paternal imprinted gene, mice with H19 deficiency from the maternal chromosome were H19−/− mice, whereas mice with H19 deficiency from the paternal chromosome were used as negative controls. Because 10-week-old WT (null) mice that underwent BDL for 7 days showed less severe liver fibrosis Supporting Fig. S1 and Fig. 1A), we used 22-week-old mice for this experiment to observe more significant differences between the control and H19−/− mice. As expected, H&E and Masson's trichrome staining revealed a marked reduction in liver injury and periductular fibrosis in H19−/− versus control mice (Fig. 5A), which was accompanied by the decreased levels of serum AST, ALT, bilirubin, and BA (Fig. 5B). Consistently, the protein (Fig. 5C) and mRNA (Fig. 5D) levels of ZEB1 were increased whereas EPCAM and SOX9 were decreased in H19−/− mice. In addition, the expression of Ck7, Ck19, Col1a1, Col1a2, and α-Sma was concomitantly reduced in H19−/− livers (Fig. 5D). However, EMT-related genes showed minimal changes in H19−/− versus control livers (Supporting Fig. S5).
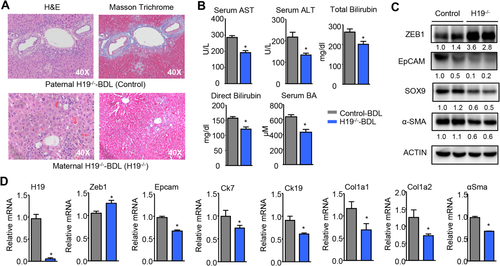
Overall, the results provided direct in vivo evidence for a regulatory role of H19 in cholestatic liver fibrosis.
BILE ACIDS INDUCED H19 IN CHOLANGIOCYTES TO CONTROL ZEB1 AND EpCAM EXPRESSION
One major phenotype in H19-BDL mice was the development of periductular fibrosis (Fig. 4A); therefore, we determined the H19/ZEB1/EpCAM regulation in cholangiocytes. We examined H19 expression regulation by various BAs in MSCs and MLCs. It was interesting to note that H19RNA was highly induced by several BAs, including CA, TCA, UDCA, and LCA (5- to 15-fold induction) in MSCs but was only modestly induced by TCA and UDCA (∼1.5 fold induction) in MLCs (Fig. 6A). However, H19RNA was only induced by LCA in primary hepatocytes of WT but not H19−/− mice (Supporting Fig. S6). Overexpression of H19 markedly reduced ZEB1 protein with a concurrent induction of EpCAM in MSCs (Fig. 6B). Interestingly, H19 showed no effect in ZEB1 expression in MLCs in which EpCAM protein could not be detected with a short exposure time.
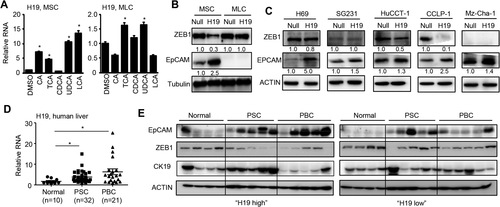
To establish the clinical relevance of H19 in cholestasis, we examined the effect of H19 in several human cholangiocyte cell lines, including human normal cholangiocyte cell line H69 and cholangiocarcinoma cell lines SG231, HuCC-T1, CCLP-1, and Mz-Cha-1. Overexpression of H19 markedly induced EpCAM with a moderate reduction of ZEB1 in H69 (Fig. 6C). In SG231, H19 moderately elevated EpCAM without affecting ZEB1 protein. In HuCCT-1, H19 moderately decreased ZEB1 with a commitment induction of EpCAM. In CCLP-1, H19 markedly diminished ZEB1 and induced EpCAM. In Mz-Cha-1 cells, H19 inhibited EpCAM although ZEB1 was not detected. The molecular basis for the differential responses of mouse and human cholangiocytes to H19 was unclear as the endogenous H19RNA was much higher in MLS and CCLP-1 (Supporting Fig. S7A,B). Nonetheless, the results suggested that H19 activation of EpCAM expression could be either dependent or independent of ZEB1 reduction in different cholangiocarcinoma cell lines.
HEPATIC H19 AND EpCAM WERE UPREGULATED IN HUMAN CHOLESTATIC LIVER FIBROSIS
PSC and PBC are the most common immune-mediated chronic cholestatic liver fibrosis. In PSC, bile ducts become blocked, leading to bile accumulation in the liver that slowly damages the bile ducts. The focal lesion typical for PSC is periductular layered fibrosis (characteristic “onion skin” pattern) seen with edema and inflammation surrounding the interlobular bile ducts.34 As shown above, a similar characteristic was observed in H19-BDL mice (Fig. 4A). PBC is a chronic autoimmune liver disease characterized by progressive destruction of the bile ducts and cholestasis. We determined H19RNA levels in normal, PSC, and PBC liver specimens and observed markedly elevated H19 expression in PSC and PBC (Fig. 6D). We divided the samples into two groups based on the relative levels of H19 versus the normal samples: H19 high and H19 low. EpCAM proteins were significantly induced in PSC in both groups but were more highly induced in PBC in H19 high versus H19 low samples (Fig. 6E). The decreased ZEB1 proteins were observed in PSC and PBC of the H19 high group compared with normal, which was negatively correlated with EpCAM expression. Interestingly, Ck19 proteins were only elevated in PSC but not PBC in the H19 high group. Taken together, these results strongly support the involvement of H19 in the pathogenesis of human cholestatic liver fibrosis.
Discussion
Albeit from different origins, including biliary obstruction, infection of hepatitis virus, alcohol consumption, and nonalcoholic steatohepatitis, liver fibrosis can progress into cirrhosis and ultimately into end-stage liver disease. Accumulating progress has been made in better understanding the underlying mechanisms of hepatic fibrosis with recent major investigation focused on the dynamic homeostasis between hepatic progenitor cells and ductular reaction, balance of epithelial cell niche and mesenchymal cell niche.35 As one of the earliest discovered lncRNAs, H19 has been recognized as a multifunctional regulator, including orchestrating the epithelial-to-mesenchymal transition (EMT) to promote tumor growth.36 In this study, we unravel a crucial function of H19 in cholestatic liver diseases. We demonstrate that obstructive cholestatic liver fibrosis is exacerbated by hepatic overexpression of H19 but is ameliorated by H19 deficiency.
We identify EpCAM as a novel downstream target gene of ZEB1. ZEB1 represses EpCAM gene transcription by the direct binding to the EpCAM promoter and H19 suppresses ZEB1 transactivation by interacting with ZEB1 to prevent its binding. Mechanistically, H19 promotes fibrosis at least in part through its activation of EpCAM by down-regulating ZEB1 in both hepatocytes and cholangiocytes. Thus, we elucidate a dual inhibitory mechanism involving H19 repression of ZEB1 and ZEB1 inhibition of EpCAM, leading to EpCAM activation by H19 (Fig. 7).
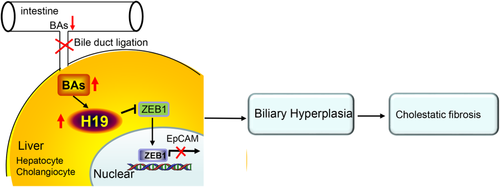
This dual inhibitory mechanism is further supported by in vivo studies. Either overexpression of ZEB1 or knockdown of EpCAM hampers H19-induced periductular fibrosis. However, EpCAM is diminished in ZEB1 mice, whereas ZEB1 remains unaltered in shEpcam mice, demonstrating that EpCAM is downstream of ZEB1 to dictate H19-mediated fibrosis. Because EpCAM is also used as a marker of biliary hyperplasia,37 the activation of EpCAM by H19 likely contributes to the ductular reaction. A recent study reported that ZEB1 is neither required nor sufficient for EMT in LS174T colorectal cancer cells lacking endogenous EMT-inducing transcription factors.38 In our studies, H19 markedly diminishes ZEB1 in MSCs that express high levels of EpCAM but has no effect on down-regulating ZEB1 in MLCs that contain low levels of EpCAM. On the other hand, H19RNA is diminished in shEpcam mice, which could be due to alleviated liver injury and decreased BA levels.
Electron microscopic studies in rodent liver sections and intrahepatic bile duct units have demonstrated that small cholangiocytes show a high nucleus-to-cytoplasm ratio and represent a hepatic progenitor cell population.34 It was reported that hepatic progenitor cells are the source of proliferating biliary epithelial cells in BDL-induced liver fibrosis.39 Consistently, H19 appeared to be more effective in small cholangicytes than in large cholangiocytes. Therefore, hepatic progenitor cells may also be involved in the pathological process of H19-induced liver fibrosis.
It is noted that in human cholangiocarcinoma cell line Mz-Cha-1, the activation of EpCAM by H19 occurs in the absence of ZEB1. H19 also induces EpCAM without affecting ZEB1 levels in SG231 cells. Therefore, H19 likely induces EpCAM via alternative mechanisms independent of ZEB1 in both cells. Impressively, the elevated EpCAM correlates with down-regulated ZEB1 expression in human PSC and PBC expressing high levels of H19, providing clinical evidence for an important role of H19 in mediating the development of biliary fibrosis.
α-SMA is one of the most reliable markers of stellate cell activation. It is increasingly used as an early indication of fibrogenic activity in human liver disease, even before extracellular matrix accumulates.22 The expression of α-SMA protein was elevated in H19-BDL mice and decreased in H19−/−-BDL mice, suggesting that the activation of hepatic stellate cells also contributed to H19-mediated fibrosis.
In conclusion, our study establishes H19 as a profibrotic mediator of cholestatic live fibrosis. The dysregulation of bile acid homeostasis results in a marked induction of H19 by bile acids in cholangiocytes and hepatocytes, and this in turn prevents ZEB1 inhibition of EpCAM, causing EpCAM activation and leading to biliary hyperplasia. Our work provides mechanistic insight into the role of lncRNA in cholestatic liver fibrosis and may pave the way for the development of therapeutic strategies to target lncRNA.
Acknowledgement
We thank Xinwei Wang (National Cancer Institute) for the Epcam-Luc construct and Meenakshisundaram Ananthanarayanan (Yale University) for cholangiocyte-derived cells.
REFERENCES
Author names in bold designate shared co-first authorship.




