Genetic variants associated with physiological and biochemical indicators: A multi-centre whole-exome sequencing study of Chinese healthy participants
IMPACT Study Group member names are provided in Acknowledgements section.
Dear Editor,
Disease onset and progression manifest through quantifiable alterations in organ function biomarkers, including hepatic, renal and lipid profiles. Even within normal reference ranges, these indicators could predict disease risk.1 Physiological and biochemical profiles in Asians display distinct characteristics compared to those in Caucasians.2 Identifying factors that influence these markers in Asian populations may therefore enhance early disease prediction and intervention, improving clinical outcomes.
Given the heritability of physiological and biochemical indicators,3 linking genetic variations with these indicators differences through genome-wide association studies (GWAS) has become a highly promising approach, particularly in lipid metabolism.4 However, databases such as UK Biobank and FinnGen present limitation due to ethnic diversity and lack of stringent screening. This study employed whole-exome sequencing to analyse genes associated with key physiological indicators in clinically screened Chinese healthy participants, providing insights into their genetic correlations.
A total of 778 participants in IMPACT study were included in this cohort.5-8 Briefly, healthy volunteers were enrolled in this study based on the absence of clinically significant abnormalities in blood pressure, heart rate, routine blood, blood biochemistry, chest X-ray, urinalysis and electrocardiogram, as well as confirmation of no infectious diseases or non-pregnancy status. Figure 1 shows the flow diagram. The physiological and biochemical indicators of participants are presented in Table 1. In discovery cohort, correlation analysis and linear regression were conducted to choose the covariates (Figure S1 and Table S1). Consequently, age, male proportion and body mass index were included as covariates in subsequent genetic association analyses. Identity-by-descent analysis excluded first-degree relatives (Proportion of identity by descent: PI_HAT ≤ .5) prior to genetic modeling.
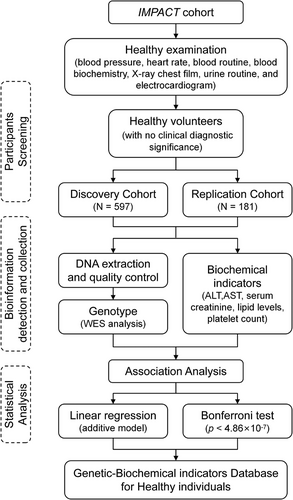
| Characteristic | Discovery cohort (N = 597), mean ± standard deviation | Replication cohort (N = 181), mean ± standard deviation | p-Value |
|---|---|---|---|
| Age | 28.71 ± 8.26 | 29.82 ± 6.65 | .064 |
| 18 ≤ age < 30 | 61.24% (371/597) | 53.04% (96/181) | |
| 30 ≤ age < 40 | 24.29% (145/597) | 35.91% (65/181) | |
| 40 ≤ age < 50 | 12.2% (73/597) | 11.05% (20/181) | |
| 50 ≤ age < 60 | 1.34% (8/597) | .00% (0/181) | |
| Male | 67.67% (193/597) | 64.64% (64/181) | .448 |
| Weight (kg) | 62.65 ± 7.63 | 62.23 ± 7.35 | .540 |
| Body mass index | 22.65 ± 1.94 | 22.40 ± 1.88 | .113 |
| Alanine aminotransferase (U/L) | 16.83 ± 9.52 | 16.65 ± 8.91 | .821 |
| Aspartate aminotransferase (U/L) | 18.47 ± 4.91 | 19.34 ± 4.58 | .034 |
| Serum creatinine (µmol/L) | 74.47 ± 16.46 | 66.80 ± 12.85 | <.001 |
| Platelet (109/L) | 254.07 ± 55.65 | Not record | No data |
| Total cholesterol (mmol/L) | 4.32 ± .73 | 4.10 ± .70 | .002 |
| Triglyceride (mmol/L) | 1.06 ± .51 | 1.13 ± .52 | .183 |
| Low-density lipoprotein (mmol/L) | 2.47 ± .62 | 2.41 ± .51 | .522 |
| High-density lipoprotein (mmol/L) | 1.37 ± .28 | 1.30 ± .33 | .144 |
To investigate genetic variants associated with liver function, genetic association analysis was conducted in total cholesterol (TC), triglyceride (TG), low-density lipoprotein (LDL), high-density lipoprotein (HDL), alanine aminotransferase (ALT) and aspartate aminotransferase (AST). The Manhattan plot of discovery cohort is shown in Figure 2. Single nucleotide polymorphisms (SNPs) correlated with liver function and disease are listed in Tables S2-S7. Notably, RBM19 rs117916372 was associated with both ALT and AST. Compared with AA carriers of rs117916372, homozygous GG carriers exhibited lower level of ALT (AA vs. GG: 16.20 ± 8.66 U/L vs. 12.75 ± 4.43 U/L, p = 1.53 × 10−7) and AST (AA vs. GG: 18.38 ± 4.61 U/L vs. 15.00 ± 4.24 U/L, p = 2.56 × 10−10). Rs3851 on TM4SF5 exhibited a correlation with ALT and AST levels in both the discovery and replication cohorts (Table S8 ).
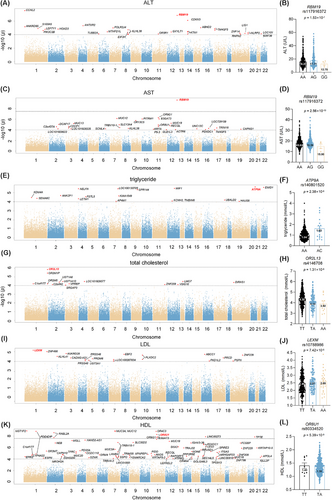
In the genome-wide analyses of TG, ATP9A rs140801520 was most significantly associated with TG levels (p = 2.38 × 10−6). UBALD2 rs712833 exhibited a correlation with TG levels in both the discovery and replication cohorts (Table S8). For TC, OR2L13 rs4146708 was primarily correlated with cholesterol variation (p = 1.31 × 10−6). For LDL, LEXM rs10788986 showed a notable association with LDL (p = 7.42 × 10−6). Regarding HDL level, multiple SNPs on OR8U1 were associated (p = 5.39 × 10−9). Notably, OR8U1 also has a potential correlation with AST level (p = 1.32 × 10−5). Compared with TT carriers of rs80334520, heterozygous TA carriers exhibited lower level of AST (TT vs. TA: 23.01 ± 9.35 U/L vs. 18.43 ± 4.63 U/L) and HDL (TT vs. TA: 1.41 ± .28 mmol/L vs. 1.38 ± .29 mmol/L). Elevated serum AST and HDL levels suggests hepatocyte damage or chronic inflammation.9 The association of OR8U1 with both AST and HDL levels indicted the potential role of olfactory receptor OR8U1 in liver metabolic-inflammatory regulation, requiring further in vivo and in vitro validation.
Genetic association analysis of serum creatinine is shown in Figure 3. SNPs correlated with serum creatinine are listed in Table S9. Among them, multiple SNPs located on NBPF1, MST1P2, NBPF10, SDHAP2, FRG1, FRG1B, MUC3A and MUC6. A mutation in rs3901679 on NBPF1 from the T to C allele was associated with a significant increase in serum creatinine levels (TT vs. TC: 72.96 ± 16.31 µmol/L vs. 86.56 ± 12.21 µmol/L). In addition, rs80068592, rs140898464 and rs61734664 exhibited a potential correlation with creatinine in both the discovery and replication cohorts, as shown in Table S8.
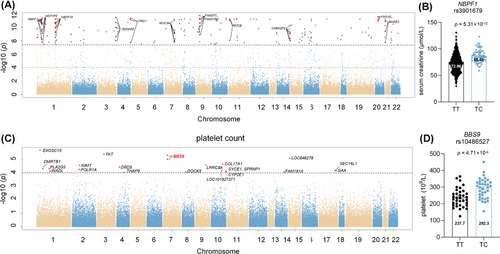
To investigate genetic variants associated with hematopoiesis, genetic association analysis was conducted with platelet count (Table S10). Several SNPs on BBS9, POLR1A and SPRNP1 genes demonstrated significant correlations with platelet count in Figure 3. Specifically, allele change from T to C on rs10486527 was associated with an increased platelet count (TT vs. TC: 244.30 ± 50.36 × 109/L vs. 292.83 ± 60.15 × 109/L, p = 4.71 × 10−6).
We also conducted cross-validation of SNPs reported in previous GWAS. No statistical significant associations was observed (Table S11). Potential significant associations were presented as follows: ZNF646 rs749671 with TG (p = .0017), TM6SF2 rs58542926 with ALT (p = .0058), DHODH rs2288002 with LDL (p = .0026) and PCNXL3 rs12801636 with TG (p = .044). Although prior findings were not fully replicated in this study, correlation analyses revealed concordant effect directions with published data. However, larger effect sizes (β and SE) were observed in this cohort, potentially attributable to its comparatively smaller sample size. Meanwhile, the inability of this study to replicate the majority of previously reported SNPs may be attributed to several factors. First, most prior cohorts included both patients and healthy controls, potentially introducing heterogeneity. Second, previous studies were predominantly conducted in European populations. Ethnic distinctions between Caucasian and East Asian groups, such as differences in minor allele frequencies, may account for the observed discrepancies.
This study has certain limitations. Baseline differences were observed between the discovery cohort and the validation cohort, potentially attributed to the limited sample size in the validation cohort. This potentially explain the inability to replicate discovery cohort results. This study did not account for participants' exercise habits or dietary patterns, which may introduce confounding effects on clinical indicators beyond genetic factors. Participants were healthy individuals with a younger average age and normal physiological and biochemical indicators compared to typical clinical patient populations. This study was conducted only among Chinese populations. Cross-ethnic comparisons study and functional validation are needed to confirm the role of the genes.
1 CONCLUSION
This study established associations between genetic variants and biochemical indicators through a database of healthy individuals. Our cohort identified genes correlated with biochemical indicators, offering promising biomarkers and therapeutic insights for future clinical disease prediction, diagnosis and drug development.
AUTHOR CONTRIBUTIONS
Conception and design: Yimin Cui and Qian Xiang. Provision of participants and study materials: Zhe Wang, Zhiyan Liu, Qiufen Xie, Guangyan Mu, Shuang Zhou, Zining Wang and IMPACT study group. Collection of data: Zhe Wang and Zhiyan Liu. Data analysis and interpretation: Zhe Wang and Qian Xiang. Manuscript writing: Zhe Wang and Qian Xiang. All the authors have read and approved the final manuscript.
ACKNOWLEDGEMENTS
We would like to thank CapitalBio Technology Co., Ltd. (Beijing, China) for their technical assistance. We would also like to thank the National High Level Hospital Clinical Research Fund (Peking University First Hospital Interdisciplinary Research Project, 2023IR06), the National Natural Science Foundation of China (82373950, 82274024, 82404749, 82404751 and 82300799), the National Key R&D Program of China (no. 2016YFC0904900) and the National High Level Hospital Clinical Research Fund (Youth Clinical Research Project of Peking University First Hospital, 2024YC04) for providing financial support. Name of the members of IMPACT Study Group are as follows: Ninghong Guo (Center of Clinical Pharmacology, The Second Affiliated Hospital of Nanchang University, Nanchang, China), Jie Huang (Center of Clinical Pharmacology, Third Xiangya Hospital, Central South University, Changsha, China), Yunlong Tan (Psychiatry Research Center, Beijing HuiLongGuan Hospital, Peking University, Beijing, China), Wenping Wang (Department of GCP Center, Affiliated Hospital of Liaoning University of TCM, Shenyang, China), Yu Cao (Office of Drug Clinical Trial Management, Affiliated Hospital of Qingdao University, Qingdao, China), Xiaohua Wei (Clinical Trial Research Center, Department of Pharmacy, The First Affiliated Hospital of Nanchang University, Nanchang, China), Yuan Li and Xin Li (Department of Pharmacy, The Third Hospital of Changsha, Changsha, China), Maoxing Liao (Department of Pharmacy, Shanghai Public Health Clinical Center, Fudan University, Shanghai, China) and Jiachun Bao (Department of Pharmacy, Affiliated Hospital of Jiangnan University, Wuxi, China).
CONFLICT OF INTEREST STATEMENT
The authors declare they have no conflicts of interest.
ETHICS STATEMENT
All studies were approved by the independent ethics committee of Peking University First Hospital and all participating centres. Subjects were informed before study and provided written informed consent. This study was registered on ClinicalTrial.org with the registration numbers NCT03161496 and NCT03161002.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are deposited in the National Population Health Data Center (https://www.ncmi.cn) and available from the corresponding author upon reasonable request (doi:10.12213/11.A0028.202009.338.V1.0).




