Single-cell RNA sequencing of peripheral blood mononuclear cells from bronchopulmonary dysplasia
Yufeng Liu, Chun Yan, Yushan Li and Ruoxing Zhou contributed equally.
Abstract
Background
Bronchopulmonary dysplasia (BPD) is a severe respiratory disease that primarily affects premature infants, characterized by persistent inflammation and abnormal immune activation. This study aimed to elucidate the immunological mechanisms underlying BPD by integrating single-cell RNA sequencing with T/B cell receptor profiling of peripheral blood mononuclear cells (PBMCs) from preterm infants with BPD, complemented by validation in a murine BPD model.
Methods
We profiled PBMCs from preterm infants diagnosed with BPD and healthy controls, identifying 22 distinct cell clusters corresponding to major immune cell types.
Results
Significant alterations were observed in myeloid and lymphoid subsets, with neutrophils undergoing metabolic reprogramming toward oxidative phosphorylation. T and B cell subsets exhibited phenotypic and functional changes, with B cells serving as crucial interaction hubs in cell communication networks. Progenitor cell analysis in BPD mouse models revealed specific alterations in hematopoietic stem cells. Analysis of cell–cell communication networks highlighted intricate intercellular interactions in BPD, emphasizing a pivotal role for the BTLA-TNFRSF14 signaling axis in disease pathogenesis. Additionally, pharmacological blockade of BTLA in mouse models alleviated disease severity, suggesting its potential therapeutic effects through modulation of the BTLA-TNFRSF14 pathway.
Conclusion
These findings enhance the understanding of the BPD immune microenvironment and lay the foundation for developing targeted immunomodulatory therapies.
Highlights
- Single-cell sequencing revealed immune cell profiles in bronchopulmonary dysplasia (BPD).
- Neutrophils underwent metabolic changes, and B cells were key in immune communication.
- Targeting B and T lymphocyte attenuator (BTLA)-TNFRSF14 signalling reduced BPD severity in mouse models, suggesting a potential therapy.
Dear Editor,
Bronchopulmonary dysplasia (BPD) is a severe respiratory disease that primarily affects premature infants, characterized by persistent inflammation and abnormal immune activation.1 This study aimed to elucidate the immunological mechanisms underlying BPD by integrating single-cell RNA sequencing (scRNA-seq) with T/B cell receptor (TCR/BCR) profiles of peripheral blood mononuclear cells (PBMCs) from preterm infants with BPD, further validated in a murine BPD model.
We used scRNA-seq to characterize PBMCs from four BPD patients and four healthy controls (Table S1), performed flow cytometry (FCM) analysis and established BPD mouse models to compare immune cell subsets in lung tissue and human peripheral blood (Figure 1A). Based on the gene signatures,2 we identified eleven major cell types, including CD4+ T cells (CD3D, CD4), CD8+ T cells (CD8A, CD8B), natural killer (NK) cells (NCAM1, KLRB1, NKG7), B cells (CD19, MS4A1), CD14+ monocytes (LYZ, CD14, CD68), CD16+ monocytes (LYZ, FCGR3A), conventional dendritic cells (cDCs; CLEC10A), plasmacytoid DCs (pDC; LILRA4),3 neutrophils (FCGR3B, CSF3R), progenitor cells (CD34) and megakaryocytes (PPBP) in the PBMCs (Figure 1B,C and Figure S1B,C). We observed a trend of decreased neutrophil abundance in BPD patients (Figure 1D), confirmed by FCM analysis of peripheral blood (Figure 1E and Figure S2A), which was also reflected in the BPD mouse model (Figure 1F and Figure S2B). To explore the causes of these immune differences, we investigated immune cell differentiation from hematopoietic stem cells (HSCs) in the bone marrow.4 FCM analysis of bone marrow samples from BPD mice revealed a significant increase in Lin−Sca1+cKit+ cells and long-term HSCs in the BPD group (Figure S3). Enrichment analysis of gene expression from all PBMCs revealed oxidative phosphorylation as the primary pathway associated with upregulated genes in BPD (Figure 1G–I).
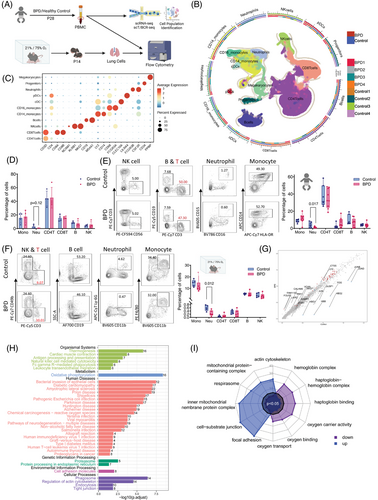
Using genes highly expressed in blood monocytes (S100A8, S100A9 and CSF3R) and classical markers (CD14, FCGR3A and LYZ),3 we classified monocytes into three subsets: CD14⁺ monocytes, CD16⁺ monocytes, and intermediate monocytes co-expressing CD14 and FCGR3A. DCs were further divided into cDCs and pDCs. Mature neutrophils (FCGR3B, CCL4 and NAMPT) and granulocytes (CAMP, LCN2 and DEFA3)5 were identified (Figure 2A,B). In BPD patients, we observed a decrease in mature neutrophils and an increase in intermediate monocytes (Figure 2C), which are known to secrete pro-inflammatory cytokines such as tumour necrosis factor (TNF)-α, interleukin (IL)-1β, and IL-6 upon stimulation.6 FCM analysis confirmed the increased secretion of these cytokines by intermediate monocytes in BPD (Figure S4). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis unveiled that the differentially expressed genes (DEGs) in mature neutrophils were predominantly implicated in the oxidative phosphorylation pathway (Figure S5A and Figure 2D), consistent with the high-energy demands of neutrophils in the injured area following mechanical ventilation.
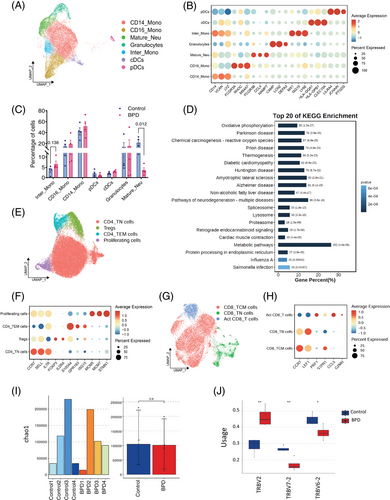
We identified seven T cell subsets. Within CD4⁺ T population, we categorized effector memory (S100A4, GPR183 and ISG15), naïve (CCR7, SELL and IL7R), regulatory T (Tregs; FOXP3 and IL2RA), and proliferating T cells (MCM5, MCM7 and STMN1)7 (Figure 2E,F). The distribution of CD4⁺ subsets was similar between BPD patients and healthy controls, with no significant differences (Figure S5B), suggesting immune dysregulation without major changes in CD4⁺ populations. Additionally, three CD8⁺ T cell subsets were identified: naïve (CCR7 and LEF1), central memory (PRF1 and S1PR1), and activated (CCL5 and GZMH)8 (Figure 2G,H). Central memory CD8⁺ T cells were slightly increased in BPD patients, indicating a potential role in infection control (Figure S5C). TCR clonal expansion analysis showed no significant differences in clonotypes or diversity between the two groups (Figure 2I and Figure S5D,E). However, changes in the distribution of V-J pairs in β chains were observed, particularly in TRBV2, TRBV6-2, and TRBV7-2 in BPD (Figure 2J). In conclusion, specific TCR clonotypes in β chains may contribute to BPD pathogenesis.
Additionally, we identified three B cell subsets (Figure S6) and five NK cell subsets (Figure S7). CellChat analysis9 revealed a comprehensive interaction network involving all subsets (Figure S8A). Notably, BPD patients exhibited more cell-to-cell interactions compared to controls, with the strength of these interactions gradually increasing, potentially driven by elevated ligand and receptor expression during disease progression (Figure 3A,B and Figure S8B). We identified 22 ligand-receptor pairs across 16 signalling pathways in BPD patients, with the GRN, CCL and B and T lymphocyte attenuator (BTLA) pathways unique to the BPD group (Figure 3C). B cells were central hubs in this network, particularly involved in BTLA signalling with T cells (Figure 3D and Figure S8C–E). The BTLA-TNFRSF14 axis emerged as a major mediator of these interactions (Figure 3E). TNFRSF14 is a membrane-bound receptor that activates nuclear factor-kappa B, and BTLA-TNFRSF14 is a well-characterized ligand-receptor pair.10 Treatment with an anti-BTLA antibody in the BPD mouse model resulted in improved alveolar development and reduced inflammatory cytokines (Figure 3F,G). TNFRSF14 levels showed a parallel modulation (Figure 3H), suggesting that inhibiting the BTLA-TNFRSF14 pathway may protect against lung damage and reduce inflammation in BPD.
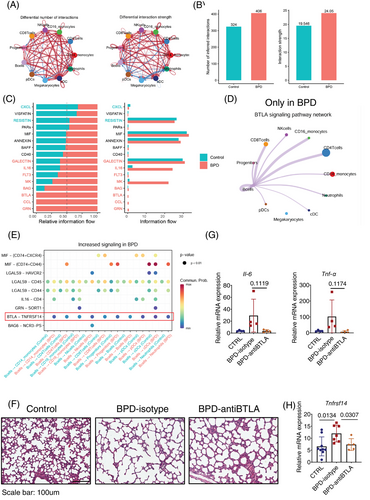
In conclusion, we found significant alterations in both myeloid and lymphoid subsets, with neutrophils undergoing metabolic reprogramming toward oxidative phosphorylation. Progenitor cell analysis in BPD mouse models revealed specific alterations in HSCs. The BTLA-TNFRSF14 signalling axis plays a key role in BPD pathogenesis, and blocking BTLA in mouse models reduces disease severity, suggesting potential therapeutic effects.
AUTHOR CONTRIBUTIONS
Yufeng Liu, Wangkai Liu, Yushan Li and Chun Yan designed experiments. Yufeng Liu, Wangkai Liu, Yushan Li, Chun Yan, Xiaoyu Lin, Qiong Meng, Sitao Li and Limei Zhong performed the experiments. Yufeng Liu, Wangkai Liu, Yushan Li, Chun Yan and Ruoxing Zhou analyzed data. Yufeng Liu, Wangkai Liu, Yushan Li, Yanfang Tan and Chun Yan wrote the manuscript.
ACKNOWLEDGEMENTS
We thank all the individuals who participated in this study.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
FUNDING INFORMATION
This work was supported by grants from National Natural Science Funds (No. 82171695); Science and Technology Program of Guangzhou (SL2024A03J01319; SL2024A04J00240); Natural Science Foundation of Guangdong Province, China (2022A1515010031); Basic and Applied Basic Research Fund of Guangdong Province (2022A1515012548).
ETHICS STATEMENT
This study was approved by the Institutional Ethics Committee for Clinical Research and Animal Trials of the First Affiliated Hospital of Sun Yat-sen University (No. [2020]521).
Open Research
DATA AVAILABILITY STATEMENT
The raw sequence data have been deposited in the Genome Sequence Archive in the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA007764). The remaining data are available from the authors upon request.




