Severe SARS-CoV-2 infection in diabetes was rescued in mice supplemented with metformin and/or αKG, and patients taking metformin, via HIF1α-IFN axis
Dear Editor,
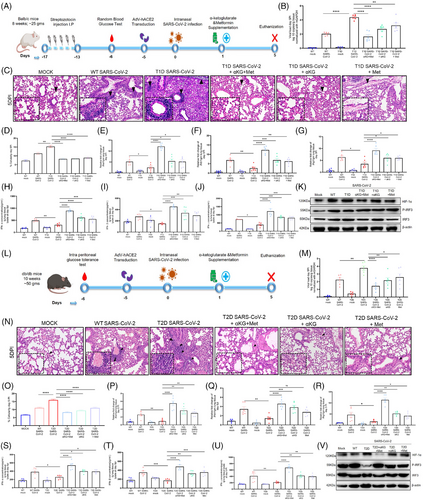
Studies report a higher prevalence of COVID-19 and worse outcomes in diabetic patients, typically 1.5-3-fold greater than similar populations without diabetes1 and the individuals taking the hypoglycaemic drug metformin have better survival.2, 3 We investigated the mechanisms for the above reports and observed that the hyperglycaemic mice with either type 1 or type 2 diabetes (T1D/T2D) have elevated SARS-CoV-2 infection in the lungs at day-5 post infection (5DPI, Figure 1 A, B) with aggressive inflammation, indicated by infiltration of immune cells (Figure 1C,D) and accumulation of cytokines TNFα and IL6 (Figure S1C,D) as compared to non-diabetic counterparts, (T2D mice data in Figure 1L–O, Figure S2C-D). Both T1D/T2D mice showed decreased levels of interferons in lung tissue lysate (type-1 and -2; IFN-α/IFN-β/IFN-γ, Figure 1H–J/S–U) alongside decreased expression of IFNA1/IFNB1/IFNG1 and IFN-regulatory factor (IRF)-3/IRF-7 genes (Figure-1E–G/P–R, Figures S1E,F and S2E,F) in the lung tissue and in the spleen (Figures S1I–M and S2I–M) but elevated HIF-1α in the lungs (Figure 1K,V and Figures S1G and S2G). The supplementation with metformin and/or alpha-ketoglutarate (αKG, previously used by us4, 5) significantly reduced viral load, and rescued inflamed lungs and IFNs synthesis (Figure-1K,V and Figure S1 H/S2 H). Unlike IFNs, the levels of IgG and the neutralization antibody titre against SARS-CoV-2 were found unaltered between WT and diabetic (T1D/T2D) mice even after metformin+αKG treatment (Figures S1N–Q and S2N,O). The repetition of above experiment in K-18 mice and high fat diet induced T2D model of K-18 showed similar observations (Figure S4A–M). In mechanism, we described that hyperglycaemic microenvironment (25 mM glucose) increased SARS-CoV-2 infection (Figure 2A) and decreased IFNA1/IFNB1 expression in the insulin resistant Huh-7 cells model (Figure S3A,B). The metformin and/or alpha-ketoglutarate treatment increased the IFN synthesis and decreased viral load (Figure 2A–C). The expression of HIF-1α inversely corelated with P-IRF3 suggesting a decreased IFN synthesis in these cells in diabetic microenvironment, and its improvement following metformin and/or αKG treatment (Figure 2D and Figure S3E,F). The inhibition of HIF-1α function by CAY10585 was found to improve IFN synthesis and inhibiting SARS-CoV-2 infection in these cells (Figure 2E and Figure S3G,H). We validated the above mechanism using HIF1α-depleted (using HIF1A-shRNA previously used by us5) U937 monocytic cells transiently expressing hACE2. HIF1A-depleted cells have a decreased viral load as compared to cells with control-shRNA (Figure 2F). Conversely, the IFNA1/IFNB1/IFNG1 transcripts were elevated in HIF1A-depleted cells than controls (Figure 2G–I). We also observed an elevated P-IRF3 in the HIF1A-depleted cells as compared to controls (Figure 2J, Figure S3M,N).
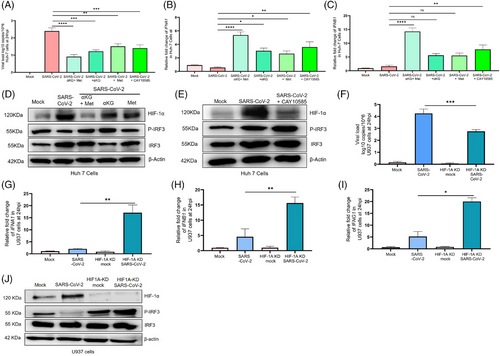
Hyperglycaemia has been associated with severe COVID-19 in patients with T1D/T2D.6, 7 Elevated glucose increases COVID-19 via HIF-1α/glycolysis pathway.6 In-vitro studies have described that hyperglycaemia increased ACE2 expression in pancreas and promotes SARS-CoV-2 infection.7 Our data have described an increased SARS-CoV-2 infection in the lungs of K-18 hACE2 mice with T2D (Figure S4). Our data also described the suppression of HIF-1α, a negative regulator of the IRF-IFN axis, by metformin in-vitro and in-vivo. HIF-1α negatively regulates IFN response. Other studies have described the correlation of low IFN levels with higher COVID-19 infection. It has been described that the hypoxic monocytes in COVID-19 patients produce less IFNα via activation of HMGB1. HIF-1α acted as a direct suppressor of IRF5/IRF3 signalling.8 Recently, we described that αKG supplementation rescued the COVID-19 pathogenesis in mice by augmenting prolyl-hydroxylase-2 (PHD2) activity, in-turn suppressing HIF-1α/P-AKT.4, 5
We further validated the above mechanisms in patients’ samples. Samples were collected between June 2020 and April 2021 when Wuhan, alpha (B.1.1.7) and delta (B.1.617.2) strains were prominent in India. The differential gene analysis of PBMCs described that the diabetic COVID-19 patients taking a higher dose of metformin (1000 mg/day) had upregulation of genes INFAR2 and IFNGR1, and pathways like interferon and cytokines in conjunction with lesser symptoms (Figure 3G, Table S1) than the counterparts taking lower dose (500 mg/day) of the drug, depicted in volcano-plot (Figure 3A), network analysis (Figure 3B), diverging bar chat (Figure 3C) and Sankey and dot-plot analysis (Figure 3D). Importantly, the above patients with high dose of metformin showed elevated levels of IFN-γ in plasma as compared to counterparts with low dose of metformin (Figure-3E). Furthermore, in-vitro stimulation of CD3-T cells from COVID-19 patients with SARS-CoV-2-RBD peptides showed an elevated IFN-γ expression in patients taking higher metformin (Figure 3F). Further, data from diabetic COVID-19 patients taking metformin revealed upregulation of genes including IFNG, IFNGR1, IFNB1, IFNA5, IRF7 and IRF3 as compared to COVID-19 patients without diabetes and without metformin (Figure 4A–C), indicating an anti-viral role of this pleiotropic drug. We validated the above findings using RT-PCR analysis. Data from diabetic COVID-19 patients taking metformin, showed higher expression of IFNA1/IFNB1/IFNG1/HIF1A and IRF-3 genes than non-diabetic COVID-19 patients without metformin (Figure 4D–H). The levels of IgG antibodies in plasma did not show any difference between diabetic COVID-19 patients with metformin and non-diabetic counterparts without metformin (Figure 4I,J) However, the neutralizing antibodies against SARS-CoV-2 were higher in metformin-treated patients than non-metformin counterparts (Figure 4K,L), indicating better adaptive immune responses of this drug against COVID-19. Several studies have described that metformin can directly impact leucocytes to suppress inflammation. Metformin significantly increased serum influenza vaccine-specific antibodies and Th-1 cell in diabetic individuals.9, 10 Likewise, we also observed an improved humoral immune response in COVID-19 patients after metformin treatment. However, a further detailed study may explain the mechanisms of our above observations. In mechanism, we observed that PBMCs of the healthy individuals treated with metformin and exposed to hypoxia (5% O2) hypoxia for 1 h showed decreased expression of HIF-1α (Figure 4M–P), indicating an inverse correlation between HIF-1α and IFN axes.
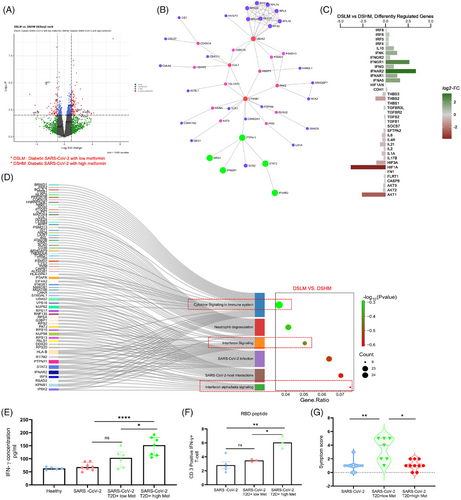

Studies together describe unique effect of metformin along with αKG in improving the IFN synthesis by suppressing HIF-1α signalling. Studies thus suggest the usage of metformin as the hypoglycaemic drug for treating diabetes on the clinical course of COVID-19. Studies also suggest that metformin in combination with αKG potentially inhibits COVID-19 (depicted in schematic Figure 4Q). This may be applicable to other viral infections that increase HIF-1α signalling and alleviate IFNs.
However, caveats of our study include the mechanism of HIF-1α-mediated alleviation of IFNs and metformin-mediated rescue in-vivo in patients. Investigating the above mechanism between diabetic COVID-19 patients without metformin or other hypoglycaemic drugs versus counterparts taking metformin. Unfortunately, samples for the earlier category are not available at our national repository.
AUTHOR CONTRIBUTIONS
Garima Joshi has designed and performed all experiments including in vivo experiments, analysed RNA sequencing data and wrote part of the original draft of manuscript. Garima Verma has performed experiments using mice and human blood samples, and wrote part of the original draft and edited the final draft of the manuscript. Simrandeep Kaur has performed in vitro and in vivo experiments. Navya Chauhan has performed PCR assays and wrote part of the original draft of manuscript. Shinjini Bhatnagar and Anil Kumar Pandey were project coordinators of the DBT India Consortium. Savita Singh and Pallavi Kshetrapal have stratified the participant samples and clinical information, and have performed clinical correlation analysis of the patients’ data. Zaigham Abbas Rizvi and Amit Awasthi performed the K-18 mice infection study and analysed data. Pradipta Jana and Bhabatosh Das have generated the RNA sequencing data, and Ellango Ramasamy and Bhabatosh Das analysed the data. Prasenjit Guchhait has conceptualized and supervised whole study and analysed data and wrote the original draft of the manuscript. All authors read, edited and approved the final manuscript.
ACKNOWLEDGEMENTS
Authors acknowledge: (1) the THSTI, NCR Biotech Science Cluster Biorepository, DBT India Consortium, for providing patients’ sample. (2) Heathy volunteers and patient participants of this study. (3) Editing by Dr. Arundhati Tiwari of the Regional Centre for Biotechnology, Faridabad, Haryana, India.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
FUNDING INFORMATION
The work was supported by the grants: BT/CS0094 from the Department of Biotechnology, Govt. of India; and ICMR/14/1923 from the Indian Council of Medical Research, Govt. of India to Prasenjit Guchhait.
ETHICS STATEMENT
Approval was also obtained from the Institutional Biosafety Committee (IBSC; ref. no. RCB/IBSC/22-23/433) of RCB. All the Covid-19-related in vitro and animal experiments were performed in the BSL3 facility of RCB. Animal experiment protocol was approved by the Institutional Animal Ethics Committee (IAEC) of Regional Centre for Biotechnology (RCB) (ref. no. RCB/IAEC/2022/116)
Open Research
DATA AVAILABILITY STATEMENT
The datasets presented in this study can be found in online repositories, GEO, NCBI, SRA, reference number SUB14556384.




