Nanopore adaptive sampling accurately detects nucleotide variants and improves the characterization of large-scale rearrangement for the diagnosis of cancer predisposition
Abstract
Background
Molecular diagnosis has become highly significant for patient management in oncology.
Methods
Here, 30 well-characterized clinical germline samples were studied with adaptive sampling to enrich the full sequence of 152 cancer predisposition genes. Sequencing was performed on Oxford Nanopore (ONT) R10.4.1 MinION flowcells with the Q20+ chemistry.
Results
In our cohort, 11 samples had large-scale rearrangements (LSR), which were all detected with ONT sequencing. In addition to perfectly detecting the locus of the LSR, we found a known MLPA amplification of exon 13 in the BRCA1 (NM_7294) gene corresponded to a duplication in tandem of both exons 12 and 13 of the reference NM_7300. Similarly, in another sample with a known total deletion of the BRCA1 gene, ONT sequencing highlighted this complete deletion was the consequence of a large deletion of almost 140 000 bp carrying over five different genes. ONT sequencing was also able to detect all pathogenic nucleotide variants present in 16 samples at low coverage. As we analyzed complete genes and more genes than with short-read sequencing, we detected novel unknown variants. We randomly selected six new variants with a coverage larger than 10× and an average quality higher than 14, and confirmed all of them by Sanger sequencing, suggesting that variants detected with ONT (coverage >10× and quality score >14) could be considered as real variants.
Conclusions
We showed that ONT adaptive sampling sequencing is suitable for the analysis of germline alterations, improves characterization of LSR, and detects single nucleotide variations even at low coverage.
Key points
- Adaptive sampling is suitable for the analysis of germline alterations.
- Improves the characterization of Large Scale Rearrangement and detects SNV at a minimum coverage of 10x.
- Allows flexibility of sequencing.
1 INTRODUCTION
The third version of sequencing generation has the potential to improve molecular diagnosis by giving a more comprehensive view of the genome. Indeed, with real-time sequencing, Oxford Nanopore Technologies (ONT) revolutionizes sequencing data generation with the sequencing of native DNA without any PCR amplification or chemical modification.1 Moreover, the real-time sequencing capability allows the specific DNA sequence selection while sequencing, without requiring previous selection during library preparation.2 This is called adaptive sampling. During sequencing of long fragments (>6000 bp), single-strand DNA molecules squeeze through protein nanopores at a speed of 400 nucleotides/second. While sequencing the first 400 nucleotides, an algorithm aligns the sequence on a reference genome and detects whether the DNA sequence corresponds to chromosomal coordinates indicated on a reference file. When the strand sequence fits with the file, the strand is fully sequenced. When the DNA strand does not correspond to the file, the sequencing stops and the DNA strand is rejected. Thus, an enrichment of specific regions is performed in real-time. Additionally, due to the sequencing data from the non-selected regions (rejected reads), a low-pass whole genome sequencing could be obtained.
For years, germline molecular diagnosis has been made increasingly accessible, and large gene panels are now routinely used in clinical practice.3 The progress of research and its application in diagnosis requires high responsiveness from routine molecular diagnosis labs. With short-read sequencing, the selection of genes is performed during library preparation by using capture probes or specific primers in multiplexed PCR. The addition of a new target needs to change the design of probe sets and different validation steps to validate the good selection of all targets. These different steps take time and have a significant cost. With adaptive sampling, an immediate responsiveness is possible and the analysis of new targets can be immediately applied to routine diagnosis. Moreover, long-read sequencing improves the detection of structural variations.4-6
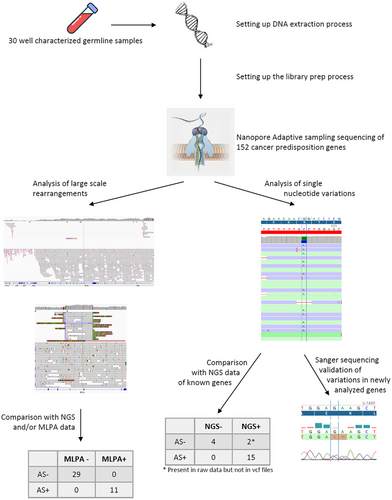
In this proof-of-concept work, we applied ONT adaptive sampling sequencing to enrich the whole sequence of 152 cancer predisposition genes to assess its capability to detect single nucleotide variations (SNV) and large-scale rearrangements (LSR) on 30 clinical samples.
2 METHODS
2.1 Patients and samples
The study was designed as a proof-of-concept study by choosing a range of alterations that are observed in a routine clinical diagnosis laboratory (Figure S1). So, we conducted it on germline blood samples from 30 patients (Table 1) diagnosed at the Georges– Francois Leclerc Cancer Center (CGFL, Dijon, France) between 2017 and 2022. These samples have representative alterations we can observe in a routine activity with techniques used in a routine molecular diagnosis lab. All germline samples were sent to our lab for analyses as part of the routine clinical diagnosis procedure for predisposing syndrome to breast, ovary, prostate, pancreas, or digestive tract cancer. All patients gave their consent to use their samples for research after their use for molecular diagnosis. The study was conducted in accordance with the Declaration of Helsinki and approved (approval no. 00010311) by the Ethics Committee of the Georges‑Francois Leclerc Cancer Center (Dijon, France) and by the Consultative Committee of Burgundy (Dijon, France) for the Protection of Persons Participating in Biomedical Research (Comité Consultatif de Protection des Personnes en Recherche Biomédicale de Bourgogne). Written informed consent was provided by all patients.
| Samples | Gene |
Mutations observed Nucleotide variation protein variation |
Classification | Clinical outcome | |
|---|---|---|---|---|---|
| #1 | BRCA1 | c.5266dup | p.(Gln1756ProfsTer74) | Pathogenic | TNBC at 43 yo |
| #2 | BRCA1 | c.(4675+1_4676-1)_(5467+1_5468-1)del | p.? | Pathogenic | Ovary cancer at 49 yo |
| #3 | MLH1 | c.1852_1854delAAG | p.(Lys618del) | Pathogenic | MSI-high colon cancer at 54 yo |
| #4 | BRCA1 | c.(134+1_135-1)_(441+1_442-1)del | p.? | Pathogenic | Ovary cancer at 68 yo |
| #5 | BRCA1 | c.(670+1_671-1)_(4185+1_4186-1)del | p.? | Pathogenic | Ovary cancer at 64 yo |
| #6 | PALB2 | c.2835-1G > C | Splicing | Pathogenic | BC at 49 yo |
| #7 | BRCA1 | c.(?_-232)_(80+1_81-1)del | p.? | Pathogenic | TNBC at 36 yo |
| #8 | BRCA1 | c.(?_-30)_(*220_?) | p.? | Pathogenic | TNBC at 33 yo |
| #9 | BRCA1 | c.4185+1_4186-1)_(4357+1_4358-1)dup | p.? | Pathogenic | TNBC at 36 yo |
| #10 | BRCA1 | c.4185+1_4186-1)_(4357+1_4358-1)dup | p.? | Pathogenic | TNBC at 37 yo |
| #11 | BRCA2 | c.2376C > A | p.(Tyr792Ter) | Pathogenic | TNBC at 49 yo |
| #12 | BRCA1 | c.5044_5048delinsT | p.(Glu1682Ter) | Pathogenic | TNBC at 57 yo |
| #13 | BRCA2 | c.5645C > A | p.(Ser1882Ter) | Pathogenic | Ovary cancer at 46 yo |
| #14 | BRCA1 | c.(5406+1_5407-1)_(5467+1_5468-1)del | p.? | Pathogenic | HR+HER2- BC at 49 yo |
| #15 | No alteration observed | TNBC at 45 yo | |||
| #16 | BRCA1 | c.1116G > A | p.(Trp372Ter) | Pathogenic | TNBC at 34 yo |
| #17 | MLH1 | c.(306+1_307-1)_(453+1_454-1)del | p.? | Pathogenic | Colon cancer at 63 yo |
| #18 | BRCA2 | c.8586_8589delinsTTCACTAAAAG | p.(Glu2863SerfsTer8) | Pathogenic | HER2+ BC at 38yo |
| #19 | MLH1 | c.2059C > T | p.(Arg687Trp) | Pathogenic | Prostate cancer at 57 yo |
| #20 | BRCA1 | c.5154G > A | p.(Trp1718Ter) | Pathogenic | Pancreas cancer at 73 yo |
| #21 | MLH1 | c.1178T > C | p.(Leu393Pro) | Pathogenic | HR+ BC at 49 yo |
| #22 | BRCA2 | c.(316+1_317-1)_(425+1_426-1)del | p.? | Pathogenic | Unaffected woman whose sister had a BC at 41 yo |
| #23 | MLH1 | c.2262del | p.(Arg755GlyfsTer28) | Unknown | Colon cancer at 33 yo |
| #24 |
BRCA1 PALB2 |
c.1016dup c.529A > T |
p.(Val340GlyfsTer6) p.(Lys177Ter) |
Pathogenic Pathogenic |
Pancreas cancer at 74 yo |
| #25 | PALB2 | c.1438A > T | p.(Lys480Ter) | Pathogenic | Bilateral TNBC at 38 yo |
| #26 | BRCA2 | c.(8487+1_8488-1)_(8632+1_8633-1)del | p.? | Pathogenic | TNBC at 33 yo |
| #27 | BRCA2 | c.5065_5066del | p.(Ala1689LysfsTer5) | Pathogenic | TNBC at 50 yo |
| #28 | No alteration observed | TNBC at 64 yo | |||
| #29 | No alteration observed | HER2+ BC at 68 yo | |||
| #30 | No alteration observed | RH+HER2- BC at 59 yo | |||
- Abbreviations: BC, breast cancer; HR, hormonal receptor; TNBC, triple-negative breast cancer; yo, years old.
2.2 DNA extraction
Three hundred microliters of whole blood were extracted with the Monarch Genomic DNA Purification Kit (New England Biolabs) by following the manufacturer's protocol. The quantity of the extracted genomic DNA was assessed with a fluorimetric method using a Qubit device (Fisher Scientific), and integrity was checked with a Tapestation 4200 device (Agilent Biotechnologies). The size of the DNA obtained was higher than 10 000 bp.
2.3 Library preparation
1.2 µg of gDNA extracted from white blood cells was slightly fragmented with g-TUBE (Covaris) by two spins of 1 min at 6010g. Then, libraries were prepared with the LSK114 kit (ONT) following the manufacturer's instructions.
2.4 Flow cell loading and sequencing set-up
R10.4.1 MinION flow cells were prepared following the manufacturer's instructions. When the final library amount was superior to 300 ng (15 µL at 20 ng/µL), half of the library was loaded. The second one was loaded 24 h later after a nuclease wash of the flow cell following the manufacturer's instructions. When the library yield is below 300 ng, the whole library is loaded at once. A bed file containing the chromosomic coordinates (±10 kb upstream and downstream of every complete gene) of 152 cancer-predisposing genes (Table S1) was uploaded to the MinKnow software, representing a size of 13 964 404 nucleotides corresponding to 0.47% of the human genome. The device used was a GridION containing a GPU Nvidia Quadro GV100.
2.5 Bioinformatics analysis
Sequenced reads were mapped using minimap2 (v2.24-r1122, -ax map-ont –MD)7 to the reference genome hg19. From the mapping, SNVs and indels of the 152 predisposing genes were identified using Clair3 (v0.1-r12, –platform = “ont” –model_path = r1041_e82_400bps_hac_g632 –enable_phasing)8 and decomposed/normalized with vt (v0.57721).9 Variants were then annotated with the following resources: VEP dbNSFP (v4.3a)10 and VEP dbscSNV (v1.1), ClinVar (2023-01),11 gnomAD (v2.1),12 OMIM (2021-12).13 Based on the annotations and ACMG guidelines,14 variants were automatically classified into five classes: pathogenic, likely pathogenic, uncertain significance, likely benign, and benign.
From the mapping, LSR were detected using Sniffles2 (v2.0.7, –phase –minsvlen 45 –long-del-coverage 10 –long-dup-coverage 0 –no-consensus).15 Variants were then annotated with the following resources: OMIM (2021-12), DGV (2016-03),16 MedGen (2022-10),17 ClinGen region and gene (2020-03),18 DECIPHER (HI_Predictions_Version3),19 and gnomAD pop frequency and pliscore (v2.1_sv.sites).20 Based on the annotations and ACMG guidelines,21 LSR were automatically classified in the 5 ACMG classes described above.
2.6 Quality score calculation
The quality score, also named the Phred score, is used to define the basecalling quality. This score is an estimated error probability. The quality score is calculated as −10log(E) where E is the estimated error probability. For example, a quality score of 20 corresponds to an error of 1 in 100.
3 RESULTS
3.1 Technical performances of adaptive sampling sequencing
We first analyzed some technical parameters to provide the best overview of this new chemistry. The first step was the library preparation yield. Indeed, even if the starting material was germline genomic DNA, some samples had been stored at −20°C for several years (mean = 1.5 years, min < 1 year, max = 5 years). The median library preparation yield was 36.98% (quantity of libraries obtained in relation to the initial quantity of gDNA). This yield was not different for samples stored for more than 2 years (median = 34.06%) or less than 2 years (38.77%; Figure 1A). Then, we tested the library preparation yield in relation to the initial quantity of gDNA. When less than 1 µg (min: 525 ng, max: 970 ng) of gDNA is used, the yield was not different from an initial quantity of 1.2 µg recommended by the provider (median = 33.09% and 38.77%, respectively). However, a higher quantity of gDNA (1.5 µg) was linked to a decrease in the library preparation yield (median = 21.35%; Figure 1B). Based on these observations, we can conclude the age of the samples did not influence the yield of the library preparation, and the optimal amount of gDNA to initiate is 1.2 µg, with an average yield of 40.86% (min: 8.86%, max: 83.08%). Consequently, even if we used the same starting amount of gDNA, we were not able to load the same amount of final library on flow cells. We therefore analyzed the sequencing throughput in relation to the amount of library loaded. The increase of the amount in fmoles but not in ng loaded on flow cells tended to improve the sequencing throughput as well as the reload of the flow cells 24 h after the first load (Figure 1C; Figure S2A). Adding up to 100 fmoles is beneficial for throughput, but more than 100 fmoles decreases throughput. Then, we correlated the number of available pores before each run with the sequencing throughput. The number of available pores at the beginning of sequencing is directly related to the sequencing throughput (Figure 1D). Finally, we also observed target gene coverage increases with the throughput (Figure 1E).
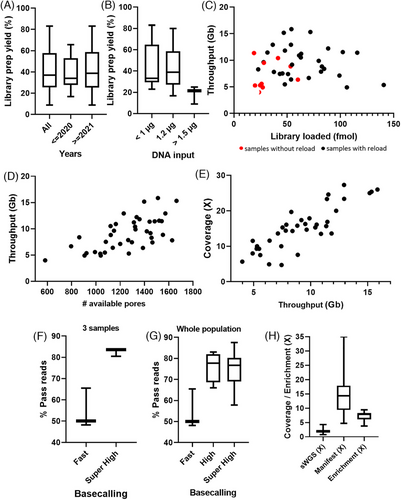
One of the specifics of ONT sequencing is the basecalling performed throughout sequencing. Three levels of basecalling accuracy were available: fast accuracy, high accuracy, and super-high accuracy. As adaptive sampling needs high informatics resources, the super-high accuracy basecalling cannot be performed simultaneously. We first tested fast-accuracy basecalling on three samples and observed only 48, 50, or 65% of the reads passed the filters. When we performed super-high accuracy basecalling on the same fast5 files (at the end of the sequencing runs), we obtained more than 80% of reads passing filters (Figure 1F). Next, we compared high-accuracy basecalling (performed during the sequencing) and super-high accuracy basecalling (performed separately from the sequencing). While the fast basecalling did not give a high percentage of good quality reads, high accuracy and super-high accuracy basecalling gave similar results with a median percentage of pass reads of about 75% (Figure 1G). Even though we observed the same results with high accuracy and super-high accuracy basecalling, we analyzed our samples with super-high accuracy once the sequencing runs had been completed. We also observed the percentage of pass reads was not influenced by the throughput (Figure S2B). Similarly, neither the percentage of reads in the target (Figure S2C) nor the enrichment (Figure S2D) were influenced by the sequencing throughput. The enrichment is directly linked to the percentage of reads in the target (Figure S2E). Moreover, enrichment had an important influence on the coverage of target genes as we obtained higher coverages with higher enrichments (Figure S2F). Finally, by using adaptive sampling, we obtained a mean coverage of shallow whole genome sequencing from the rejected gene of 2.04× (0.76–4.29), whereas the mean coverage on the genes contained in the gene selection file (also called manifest) was 14.9× (4.71–35.08), corresponding to a mean enrichment of 7.26× (3.74–9.42; Figure 1H; Figure S2G).
3.2 ONT adaptive sampling improves the characterization of LSR
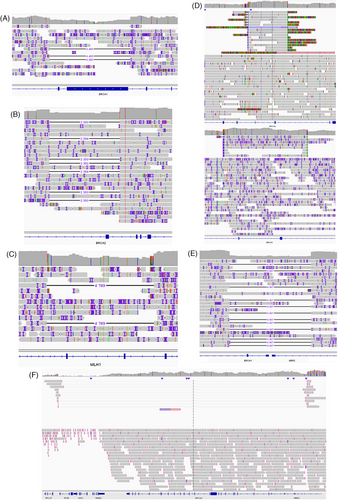
Among our 30 samples, 11 carried an LSR on different genes detected by MLPA (Multiplex Ligation-dependent Probe Amplification; Table 2). As ONT sequencing allows the analysis of long DNA fragments, we wondered whether the sequencing of complete genes from a bed file could detect the same LSR as MLPA. As expected, all LSR were also detected with ONT adaptive sampling (Table 2). Moreover, with the sequencing of introns, we were also able to identify the exact start and stop coordinates of each LSR (Table 2), and the exact size of deletions or amplifications. For example, sample #5 harbored a deletion of exons 11 and 12 of BRCA1 by MLPA (Figure S3A). With adaptive sampling, we identified a deletion of 4483 bp from intron 10 to intron 12 (Figure 2A, Table 2). We also perfectly characterized two other deletions of exon 4 in BRCA2 (Figure S3B) and exons 4 and 5 in MLH1 (Figure S3C). Indeed, we observed a 2393 bp deletion, from intron 3 to intron 4, carrying exon 4 of BRCA2 (Figure 2B, Table 2), and a deletion of 4789 bp deletion from intron 3 to intron 5, carrying exons 4 and 5 of MLH1 (Figure 2C, Table 2). In addition to identifying some well-known LSR, we also better characterized some others. Indeed, a sample originally identified as having an exon 13 duplication in BRCA1 (NM_7294; Figure S3D up), was characterized as a carrier of duplication of both exons 12 and 13 of BRCA1 (NM_7300; Figure 2D up, Table 2). We confirmed this observation on another sample carrying the same alteration (Figure 2D down, Figure S3D down). Another sample was identified with a partial deletion of promoter and a complete deletion of exon 1a and exon 2 of BRCA1 (Figure S3E). ONT sequencing showed a large deletion of 4967 bp carrying whole exons 1 and 2 of BRCA1 but also exon 1 of the non-coding protein NBR2 gene (Figure 2E). Finally, we considered another sample harbouring a complete deletion of BRCA1 (Supplemental_Fig_S3F). We confirmed the complete deletion of the BRCA1 but observed this deletion was part of a larger deletion of 139 603 bp carrying over five complete genes NBR2, BRCA1, RND2, VAT, and IFI35 (Figure 2F).
| Samples | Gene | Exons | Rearrangements observed with MLPA | Rearrangements observed with nanopore | Start | Stop | Size (bp) |
|---|---|---|---|---|---|---|---|
| #2 | BRCA1 | 16, 17, 18, 19, 20, 21, 22, 23 | c.(4675+1_4676-1)_(5467+1_5468-1)del | c.4676-1720_5467+1434del | Chr17: 41 198 226 | Chr17: 41 224 975 | 26 749 |
| #4 | BRCA1 | 5, 6, 7 | c.(134+1_135-1)_(441+1_442-1)del | c.134-530_441+1760del | Chr17: 41 254 380 | Chr17: 41 259 079 | 4700 |
| #5 | BRCA1 | 11, 12 | c.(670+1_671-1)_(4185+1_4186-1)del | c.671-115_4185+447del | Chr17: 41 242 514 | Chr17: 41 246 992 | 4479 |
| #7 | BRCA1 | 1, 2 | c.(?_-232)_(80+1_81-1)del | c.1-3845_80+1042del | Chr17: 41 274 992 | Chr17: 41 279 958 | 4967 |
| #8 | BRCA1 | All | c.(?_-30)_(*220_?) | c.1-21476_5592+39708del | Chr17: 41 157 987 | Chr17: 41 297 589 | 139 603 |
| #9 | BRCA1 | 13 | c.(4185+1_4186-1)_(4357+1_4358-1)dup | c.4186-1879_4357+4217dup | Chr17: 41 230 204 | Chr17: 41 236 471 | 6268 |
| #10 | BRCA1 | 13 | c.(4185+1_4186-1)_(4357+1_4358-1)dup | c.4186-1879_4357+4217dup | Chr17: 41 230 204 | Chr17: 41 236 471 | 6268 |
| #14 | BRCA1 | 23 | c.(5406+1_5407-1)_(5467+1_5468-1)del | c.5470-754_5530+1437del | Chr17: 41 198 193 | Chr17: 41 200 474 | 2282 |
| #17 | MLH1 | 4, 5 | c.(306+1_307-1)_(453+1_454-1)del | c.307-856_453+1270del | Chr3: 37 045 036 | Chr3: 37 049 824 | 4789 |
| #22 | BRCA2 | 4 | c.(316+1_317-1)_(425+1_426-1)del | c.317-1920_425+364del | Chr13: 32 897 293 | Chr13: 32 899 685 | 2393 |
| #26 | BRCA2 | 20 | c.(8487+1_8488-1)_(8632+1_8633-1)del | c.8488-170_8632+66del | Chr13: 32 944 923 | Chr13: 32 945 303 | 381 |
3.3 ONT adaptive sampling accurately detects single nucleotide variants
With the previous chemistry of ONT sequencing, the detection of SNV showed a high error rate. In order to test the accuracy of the new chemistry, we analyzed SNV detection on 15 samples with likely pathogenic/pathogenic (n = 15) or unknown (n = 2) variants previously detected by short-read sequencing and 4 samples without any variants detected by short-read sequencing (Table 3). All pathogenic variants except two were listed in the vcf files obtained after the alignment and variant calling. By focusing on the alignment files (bam format) for the two undetected variants, we observed them directly with an allelic frequency between 25 and 40% (Table 3). The two variants were not bioinformatically detected, probably due to their nucleotide environment and the sequence complexity created by the mutations. Indeed, homopolymer regions and highly repeated regions are difficult to sequence. Despite important advances in chemistry, sequencing could create artefacts, whatever the technology. For that, it is important to be able to discriminate true from false SNV. With short read sequencing, minimum coverage, and quality scores are now well established to validate variants.22 With ONT sequencing, especially with adaptive sampling and sequencing of native DNA, these quality criteria are not established yet. That is why we performed some analyses to find the minimum coverage and quality score allowing confidence in SNV detection. Ten samples were sequenced multiple times (2 or 3 times) due to low sequencing throughput. We performed variant analysis on the 21 runs and the 14 concatenated files obtained from the different runs. For each sample, we selected a specific SNV and followed its coverage and quality score for each run and each concatenation, representing a total of 35 SNV. We know the improvement of sequencing throughput increases coverage. Herein, we observed increase in coverage tended to improve the quality score of SNV for a majority of samples (Figure 3A). Moreover, we observed a main the part of the dots had a quality higher than 14. Then, we applied this quality threshold to the variants detected with SeqOne's bioinformatics pipeline and listed in Table 3. Eleven of 13 (85%) variants presented a quality higher than 14 (Figure 3B). The SNV of the 2 variants with a lower quality score were insertions. Nucleotide insertion or deletion-induced sequencing or bioinformatics issues as both variations had quality scores below 14. These variants, not detected with our bioinformatics pipeline, were all frameshift mutations.
| Samples | Gene | Mutations observed | Allele frequency/read depth with short read sequencing | Allele frequency/read depth with nanopore sequencing | Classification | Quality score | |
|---|---|---|---|---|---|---|---|
| #1 | BRCA1 | c.5266dup | p.(Gln1756ProfsTer74) | 43.4%/491 | 54.5%/11 | Pathogenic | 9 |
| #3 | MLH1 | c.1852_1854delAAG | p.(Lys618del) | 51.6%/184 | 40%a/10 | Pathogenic | Not available |
| #6 | PALB2 | c.2835-1G > C | Splicing | 45.1%/91 | 21.4%/14 | Pathogenic | 16.41 |
| #11 | BRCA2 | c.2376C > A | p.(Tyr792Ter) | 48.7%/1088 | 50%/30 | Pathogenic | 42 |
| #12 | BRCA1 | c.5044_5048delinsT | p.(Glu1682Ter) | 44.4%/1912 | 80%/10 | Pathogenic | 15 |
| #13 | BRCA2 | c.5645C > A | p.(Ser1882Ter) | 48.7%/3149 | 53.8%/18 | Pathogenic | 20 |
| #15 | No alteration observed | ||||||
| #16 | BRCA1 | c.1116G > A | p.(Trp372Ter) | 46.8%/1589 | 60%/20 | Pathogenic | 24.94 |
| #18 | BRCA2 | c.8586_8589delinsTTCACTAAAAG | p.(Glu2863SerfsTer8) | 35.5%/1112 | 33.3%/21 | Pathogenic | 11.29 |
| #19 | MLH1 | c.2059C > T | p.(Arg687Trp) | 52.4%/338 | 28.6%/37 | Pathogenic | 27 |
| #20 | BRCA1 | c.5154G > A | p.(Trp1718Ter) | 42.9%/655 | 53.8%/13 | Pathogenic | 25 |
| #21 | MLH1 | c.1178T > C | p.(Leu393Pro) | 48.8%/863 | 44%/9 | Pathogenic | 21 |
| #23 | MLH1 | c.2262del | p.(Arg755GlyfsTer28) | 44.3%/1434 | 57.1%/7 | Unknown | 21 |
| #24 |
BRCA1 PALB2 |
c.1016dup c.529A > T |
p.(Val340GlyfsTer6) p.(Lys177Ter) |
46.6%/223 54.1%/109 |
25%a/24 51.9%/27 |
Pathogenic Pathogenic |
Not available 28 |
| #25 | PALB2 | c.1438A > T | p.(Lys480Ter) | 49.6%/964 | 36.4%/22 | Pathogenic | 25 |
| #27 | BRCA2 | c.5065_5066del | p.(Ala1689LysfsTer5) | 35.8%/542 | 42.4%/13 | Unknown | 23 |
| #28 | No alteration observed | ||||||
| #29 | No alteration observed | ||||||
| #30 | No alteration observed | ||||||
- a Single nucleotide variations only present in raw data.
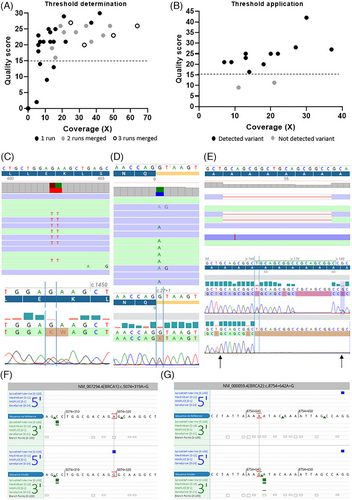
Quality score threshold is not sufficient to validate a mutation and guidelines need to apply a minimum read depth. With short read sequencing, the minimum read depth is 30×. With ONT adaptive sequencing, native gDNA is sequenced without any PCR amplification. This specificity completely abolishes the appearance of PCR artefacts. Based on this specificity, we thought a read depth lower than 30× could be applied. We observed a majority of good-quality mutations had a coverage higher than 10× (Figure 3A). By applying these filters (quality > 14 and coverage > 10×), we were able to detect 82.9% (Figure 3B) of already known variants present in Table 3.
Then, we tested if we could confirm, with Sanger sequencing, variants selected by applying both thresholds on genes that were not analyzed by short-read sequencing. We randomly selected six variants in six different genes (Table S2) representing different types of alterations (small deletion, transition, transversion, double point mutations) for which we designed specific PCR primers for Sanger sequencing. All variants were confirmed by the gold standard: the double mutation c.703_704delinsAA in the GPT gene (Figure 3C), the splicing mutation c.27+1G > T in the CFAP126 gene (Figure 3D), the deletion c.162_179del in the MSH3 gene (Figure 3E), and three other point mutations in ANKRD26, TFKC, and BLM genes (Figure S4).
Finally, the use of adaptive sampling allows the analysis of whole gene sequences and therefore introns. In a sample with a family cancer history but without any alteration in coding sequences of cancer-predisposing genes analyzed by short-read sequencing, we observed some deep intron variants meeting our quality and coverage criteria. After checking in silico the splicing impact of these variants, we observed that two of them could create new alternative splicing sites in either BRCA1 with a new 5′ splicing site (Figure 3F) or BRCA2 with a new 3′ splicing site (Figure 3G). These variations need to be characterized with specific methods as minigene experiments. Unfortunately, we were not able to study their impact as it was impossible to obtain new blood samples dedicated to RNA analysis from these patients.
4 DISCUSSION
In this work, we validated the technical performances of adaptive sampling from germline DNA samples. We set up the amount of gDNA to reach the best yield of library preparation. We showed age of the DNA did not impact library preparation yield, and sequencing throughput was directly impacted by the number of available pores before starting sequencing. Finally, the use of high accuracy or super-high accuracy basecalling was mandatory to obtain a higher enrichment of targets. In this way, we showed that the limitations and drawbacks discussed in the work from Loose et al.2 have been overcome with the R10.4.1 MinION flowcells with the Q20+ chemistry on a GridION device. Indeed, within the originator publication, the authors pointed out that sequencing and alignment speeds needed to be improved. With the new version of the MINknow control software and the GridION calculation capacities associated with the new R10 flow cells, the supplier has announced the decision time is below 1 s corresponding to 400 sequenced nucleotides.
Long-read sequencing is well known for giving a high resolution for the detection of structural variations but, only one case report showed the usefulness of adaptive sampling on BRCA1 gene.23 Among the 30 samples we studied, we were able to perfectly determine the start and stop coordinates of each LSR. This exact determination allowed us to show that a duplication of exon 13 (reference NM_7294) in BRCA1, detected with MLPA, was a duplication in tandem of both exons 12 and 13 (reference NM_7300). For another sample that carried a complete BRCA1 gene deletion, we highlighted this deletion was part of a larger deletion of 139,603 bp carrying over 4 other genes: NBR2, RND2, VAT, and IFI35. The knowledge of this specific large deletion gave a new look at cancer risk. Indeed, the NBR2 gene encodes a long noncoding RNA that suppresses tumour development through regulation of adenosine monophosphate-activated protein kinase activation.24 Another sample had a large deletion of the promoter and both exons 1 and 2 of BRCA1 gene, also carrying the promoter and the first exon of NBR2 gene. Consequently, the loss of both BRCA1 and NBR2 genes could increase the risk of cancer development and impact the clinical follow-up. Based on our observations, long read adaptive sampling has a better resolution than MLPA which is largely used in molecular diagnosis labs because of its low cost. Adaptive sampling has the same resolution as whole genome sequencing, which is the gold standard. Consequently, adaptive sampling could be a better tool than MLPA for the detection and characterization of LSR in routine molecular diagnosis labs, as it has the same resolution as whole genome sequencing, but at a lower cost.
The most frequent alteration in cancer-predisposing genes is SNV. A case report describing the comparison of short-read and long-read adaptive sampling showed a perfect concordance for a pathogenic duplication in the RB1 gene between both technologies.25 Herein, we tested 19 samples previously analyzed with a 35-gene panel by short-read sequencing, representing 16 well-known alterations. We confirmed, on raw data (bam files), that all SNV were present after ONT sequencing, but two of these were not listed after applying automatic variant calling. This was due to the high complexity of these SNV that were either present in highly repeated regions or induced mononucleotidic repeats, making the sequence difficult to analyze. Consequently, both basecalling and bioinformatics pipelines need to be improved.
We also tested whether SNV detected on genes not previously analyzed could be confirmed by Sanger sequencing. By applying filters based on a quality score higher than 14, and a coverage higher than 10×, we confirmed all selected SNV (three different point mutations, two consecutive point mutations, and a small deletion) with Sanger sequencing. This is the first time that SNV detected by long-read adaptive sampling was confirmed by Sanger sequencing. This suggests that, with specific coverage (>10×) and quality (>14) filters, we can be confident in SNV detected from native DNA by ONT sequencing. Adaptive sampling selects genomic regions throughout the sequencing process, and, contrary to capture-based enrichment used with short-read sequencing, the whole sequence of genes is sequenced. This highlighted a high number of unknown significance variants as deep intron variants have not been studied routinely yet. Indeed, with short-read sequencing, the only way to catch these deep intron variations is whole genome sequencing26, 27 and/or whole transcriptome sequencing28, 29 that are not currently used in routine molecular diagnosis laboratories. Although these variants are deeply intronic and not currently sought, they may be pathogenic.30, 31 Adaptive sampling could be a good alternative between short-read panel and whole genome or transcriptome sequencing. As an example, one of our samples without known alteration but with a family cancer history, had two deep intron variants predicted, in silico, as creating new alternative splicing sites in BRCA1 (new 5′ splicing site), and BRCA2 (new 3′ splicing site). Further analyses should be performed to determine whether these new splicing sites could impact the splicing of BRCA1 or BRCA2 mRNA, for example with minigene experiments, but the systematic analysis of non-coding sequences of cancer-predisposing genes could improve the efficiency of molecular diagnosis.
Herein, we only focused on LSR and SNV, but ONT sequencing can also detect methylation of cytosine from native DNA.32 Even if methylation status is not usually studied routinely, some methylation marks can be inherited and play a role in cancer predisposition.33, 34 These marks are not necessarily located in promoters of predisposing genes and therefore could be missed. However, adaptive sampling selects regions of interest throughout sequencing. From rejected reads giving a low pass whole genome sequencing, methylation or other parameters can be studied as genome-wide SNP, or more specifically SNP involved in a polygenic risk score.35-38
In conclusion, we showed that the ONT adaptive sampling sequencing workflow was easy to manage (Figure S5) and is suitable for the analysis of germline alterations. It improves the characterization of LSR and is able to detect SNV even at low coverage (>10×; Figure 4). The multiplexing of samples on PromethION flow cells will increase the coverage, improve the quality of variant detection, and reduce the cost of the analysis. Moreover, the flexibility of ONT sequencing (from a single sample to numerous samples at the same time) would improve the turn-around time. Nevertheless, an improvement in the detection of variants located in homopolymer regions should be considered. Once such an improvement is done, larger comparison studies between adaptive sampling and short read sequencing should be performed to obtain performance data (sensitivity, specificity, positive predicting value, negative predicting value…), and, then to adapt international guidelines to third-generation sequencing for germline diagnosis.
AUTHOR CONTRIBUTIONS
Study design and supervision: Romain Boidot. Data curation: Romain Boidot. Investigation: Sandy Chevrier. Result analysis: Romain Boidot, Corentin Richard, Marie Mille, and Denis Bertrand. Result visualization: Romain Boidot. Manuscript writing: Romain Boidot.
ACKNOWLEDGEMENTS
The authors thank Dr. Pratik Singh for his precious help throughout the project and for the review of the manuscript and Dr. Michael Blum for the review of the manuscript. The cost of reagents was supported by ONT.
CONFLICT OF INTEREST STATEMENT
The cost of reagents was supported by ONT. MM and DB are employees of SeqOne Genomics. RB is a paid consultant of SeqOne Genomics.
ETHICS STATEMENT AND CONSENT TO PARTICIPATE
All patients signed an informed consent to participate in the trial. The trial protocol was approved by an institutional review committee and by the relevant French regional ethical committee (Comité de protection des personnes Est).
CONSENT FOR PUBLICATION
All patients, by giving their consent to participate, have given their consent for publication.
Open Research
DATA AVAILABILITY STATEMENT
Genomic data may be shared upon reasonable request to the corresponding author in accordance with the French law on genomic data.




