Deregulation of apolipoprotein C2 gene in cancer: A potential metabolic vulnerability
Abbreviations
-
- AML
-
- acute myeloid leukemia
-
- APOC2
-
- apolipoprotein C2
-
- FA
-
- fatty acid
-
- OS
-
- overall survival
Dear Editor,
Apolipoprotein C2 (APOC2), an activator of lipoprotein lipase, participates in hydrolysis of triglycerides, very low-density lipoproteins, and high-density lipoproteins to release free fatty acids (FA).1 FA oxidation has emerged as an important source of energy for cancer survival and growth.2 Patterns of APOC2 genomics and transcriptomic alterations in cancer remained unexplored. Here, we characterize APOC2 deregulation in cancer by analyzing 176 studies (supplementary-methods).
In 46706 samples of 34 different cancers (Table S1), amplification, mutation, and deep deletion were the main identified APOC2 alterations (Figure S1A). Approximately 1% (251 patients) had at least one genetic alteration in APOC2 with the highest frequency (9.72%) present in bladder cancer and gene amplification being the most common alteration. The highest frequency of deep deletions in APOC2 occurred in diffuse glioma (1.36%). APOC2 mutations occurred in 2.3% of non-melanoma skin cancer and at lower frequencies in other cancers (Table S2). Among the 26 different APOC2 missense and truncating mutations, two were previously reported in hypertriglyceridemia (Figure S1B; Tables S3 and S4).
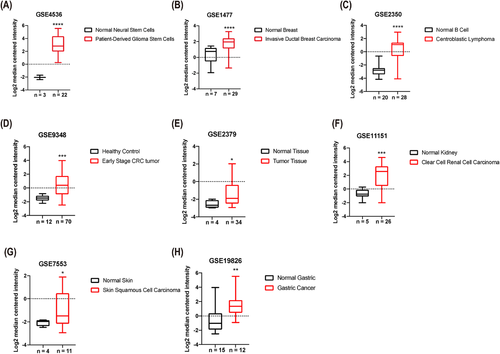
APOC2 was expressed at significantly higher levels in several malignancies (Figures 1 and S2), such as patient-derived glioblastoma stem cells (33.7-fold, p < 0.0001; Figure 1A), invasive ductal breast cancer (2.9-fold, p < 0.0001; Figure 1B), centroblastic lymphoma (10.1-fold, p < 0.0001; Figure 1C), early stage colorectal tumor (3.8-fold, p = 0.0001; Figure 1D), hypopharyngeal cancer (2.1-fold, p = 0.04; Figure 1E), clear cell renal cell carcinoma (5.9-fold, p < 0.001; Figure 1F), skin squamous cell carcinoma (2.3-fold, p = 0.023; Figure 1G), and gastric cancer (4.8-fold, p = 0.002; Figure 1H) compared with the corresponding normal tissues.
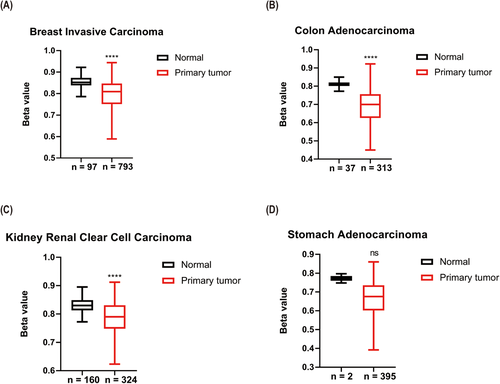
We also compared APOC2 DNA methylation beta (β) values corresponding to APOC2 gene between primary tumors and normal tissues. APOC2 median β-values were significantly lower in tumor samples compared with controls (breast invasive carcinoma: 0.809 vs. 0.852; p < 0.0001; Figure 2A; colon adenocarcinoma: 0.7 vs. 0.81; p < 0.0001; Figure 2B; kidney renal clear cell carcinoma: 0.79 vs. 0.83; p < 0.0001; Figure 2C; and stomach adenocarcinoma: 0.676 vs. 0.772; p = 0.116; Figure 2D).
The association between APOC2 alteration and chromosomal alterations, clinical attributes, and mutational status are summarized in Figure S3 and Table S5. APOC2 gene alterations were more frequently found in patients with TP53, RYR1, RYR2, and LRP1B mutations (p < 0.01; Table S6).
We further evaluated cell signaling pathways associated with APOC2 genetic alterations (amplification, mutation, and high mRNA levels) by ingenuity pathway analysis in several solid malignancies. Several pathways involved in lipid metabolism were altered (the upregulation of phospholipase C signaling and the downregulation of PPAR signaling), and pathways involved in innate and adaptive immune response were also modulated (Figure S4). The dendritic cells maturation pathway, pathways involved in the cross talk between dendritic cells and natural killer cells, Th1 and Th2 activation pathway were upregulated, while the PD-1 and PD-L1 cancer immunotherapy pathway was downregulated (Tables S8 and S9).
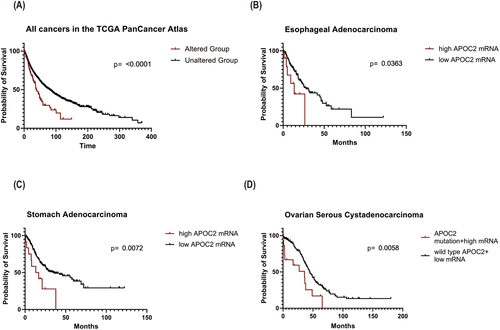
Survival analysis on all patients from different datasets showed that patients with APOC2 alterations (mutations and copy number alterations) had significantly shorter median overall survival (OS) compared with patients without alterations (OS: 37.97 vs. 80.68 months; p < 0.0001; Figure 3A). Association between altered APOC2 and poor OS was also observed in several types of cancers (Table S10). In esophageal adenocarcinoma, patients with high APOC2 expression exhibited shorter OS (13.48 vs. 28.11 months; p = 0.0363; Figure 3B). In stomach adenocarcinoma, patients with high APOC2 had a significantly shorter OS (14.96 vs. 36.00 months; p = 0.0072; Figure 3C) than patients with low APOC2. In ovarian serous cystadenocarcinoma, patients with APOC2 mutation or high expression had significantly poorer OS than patients with wild-type APOC2 and low gene expression (35.80 vs. 44.51 months; p = 0.0058; Figure 3D).
Metabolic reprogramming is a hallmark of cancer cells to support tumorigenesis and tumor progression. Lipid metabolism is elevated in cancer to satisfy increased energy demands.2 Considering APOC2's known function in FA transport and metabolism, its upregulation is a plausible mechanism for cancer adaptation and growth. By analyzing public genomics data, we found that APOC2 was upregulated and hypomethylated in several types of cancers. We recently reported APOC2 upregulation and hypomethylation in acute myeloid leukemia (AML) and more notably in AML with mixed-lineage leukemia (MLL)-rearrangements suggesting an epigenetic mechanism regulating APOC2 expression.3 Additionally, the transcription factor STAT1 interacts with RXRα on APOC2 promoter to drive APOC2 expression in macrophages.4 Upregulation of APOC2 in gastrointestinal stromal tumor was associated with cancer cell proliferation, migration, and invasion.5 Targeting APOC2 in AML caused antileukemia activity via inhibiting CD36-ERK pathway.3
Consistent with our findings, high APOC2 serum levels were previously found to be associated with shorter survival in pancreatic cancer.6 Upregulation of APOC2 was also found in metastatic tumors such as breast invasive carcinoma and fast-growing lymphoma, suggesting that APOC2 may serve as a prognostic biomarker in cancer.
APOC2 deregulations are associated with TP53 mutations. TP53 mutations are enriched in early clonal stages regardless of tumor type.7 Whether APOC2 upregulation is also an early event in tumor evolution remains to be determined. P53 is also involved in metabolic mechanisms contributing to tumor suppression.8 P53 binding to the promoter region of SREBP-1 represses the expression of SREBP-1, leading to downregulation of enzymes involved in FA synthesis.8 The mechanistic link between P53 mutations and APOC2 deregulation remains to be elucidated. APOC2 deregulations are also associated with mutations in LRP1B, a low-density lipoprotein receptor, and tumor suppressor.9
The association between APOC2 alterations and the deregulation of immune-related signaling pathways is particularly interesting. As a secreted protein that binds to CD36 receptor, APOC2 may contribute to cancer cells interaction with the immune microenvironment. CD36 mediates the metabolic adaptation which supports regulatory T-cell survival in tumors.10 Whether APOC2 contributes to the metabolic adaptation of intratumoral immune cells remains unknown.
APOC2 gene alterations and upregulation occur frequently in cancer. Functional and mechanistic studies establishing APOC2 as a therapeutic target and expanding these analyses to other apolipoprotein family members are warranted.
ACKNOWLEDGMENTS
We would like to thank the Bioinformatics core at the Norris Medical Library, University of Southern California and TCGA. We acknowledge the University of Southern California School of Pharmacy Seed Fund, the Norris Cancer Center pilot funding, and the Ming Hsieh foundation grants for HA.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
AUTHOR CONTRIBUTIONS
Yuqiao Liu performed the analyses and generated the figures. Yiting Meng performed and wrote the expression analysis. Tian Zhang performed the survival analysis. Yuqiao Liu and Houda Alachkar wrote the manuscript. Houda Alachkar conceived the project, designed the research, and supervised the project.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study were derived from the following resources available in the public domain:
cBioPortal (https://www.cbioportal.org/)
Oncomine database (https://www.oncomine.org/resource/main.html)
NCBI-GEO database (https://www.ncbi.nlm.nih.gov/gds)
UALCAN database (http://ualcan.path.uab.edu/analysis.html)




