Bromelain inhibits SARS-CoV-2 infection via targeting ACE-2, TMPRSS2, and spike protein
The new coronavirus, SARS-CoV-2, transmits rapidly from human-to-human resulting in the ongoing pandemic. SARS-CoV-2 infects angiotensin-converting enzyme 2 (ACE-2) expressing lung, heart, kidney, intestine, gall bladder, and testicular tissues of patients, leading to organ failure and sometimes death.1, 2 Currently, COVID-19 patients are treated with different agents, including favilavir, remdesivir, chloroquine, hydroxychloroquine, lopinavir, darunavir, and tocilizumab.3, 4 However, the safety and efficacy of those drugs against COVID-19 still need further confirmation by randomized clinical trials. Hence, there is an emergent need to repurpose the existing drugs or develop new virus-based and host-based antivirals against SARS-CoV-2. Bromelain is a cysteine protease isolated from pineapple stem and is used as a dietary supplement for treating patients with pain, inflammation,5 thrombosis,6 and cancer.7
Recently, studies have shown that SARS-CoV-2 homotrimeric viral spike protein (S1) binds to the Transmembrane Serine Protease 2 (TMPRSS2) primed host cell's receptor ACE-2 for initial entry, followed by S2-mediated membrane fusion.8 Of several normal and cancerous cells tested, VeroE6 and Calu-3 cells showed ACE-2 protein expression (Fig. 1A), as well as a basal level of TMPRSS2 protein (Fig. 1B). Since ACE-29 and TMPRSS2 (UniProtKB-O15393) contains cysteine residues with disulfide bonds to stabilize the protein structure, we investigated the effect of bromelain on ACE-2 and TMPRSS2 expression. Bromelain-induced a dose- and time-dependent reduction of ACE-2 and TMPRSS2 expression in VeroE6 cells (Fig. 1C and D) but do not alter ACE-2 expression in Calu-3 cells (Fig. 1E). However, bromelain reduces the expression of TMPRSS2 in Calu-3 (Fig. 1E) and ACE-2 negative normal bronchial epithelial (BEAS-2B) and lung adenocarcinoma (A549) cells (Fig. 1F and G). Cysteine protease inhibitor (E-64) treatment further confirmed that bromelain's cysteine protease activity could cleave/reduce the expression of ACE-2 and TMPRSS2 (Fig. 1H). Surface plasmon resonance (SPR) analysis revealed that purified SARS-CoV-2 S-ectodomain binds with ACE-2 in a concentration-dependent manner and has a comparable binding affinity as control RBD (Fig. 1I). The calculated molecular weight of the purified S-ectodomain-GFP protein is ∼165 kDa; however, we observed a higher molecular weight of S-ectodomain (∼215 kDa), which may be due to heavy N- and O-linked glycosylation (Fig. 1I intent). A serological assay showed a significantly increased median fluorescent intensity (MFI) of purified S-ectodomain with COVID-19 positive patients’ samples (Fig. 1J). These two results indicated that purified S-ectodomain is a properly folded and functionally active protein.
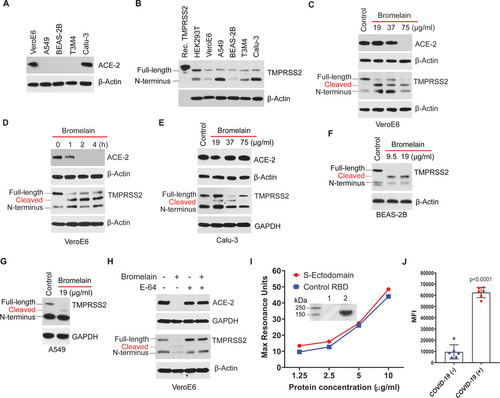
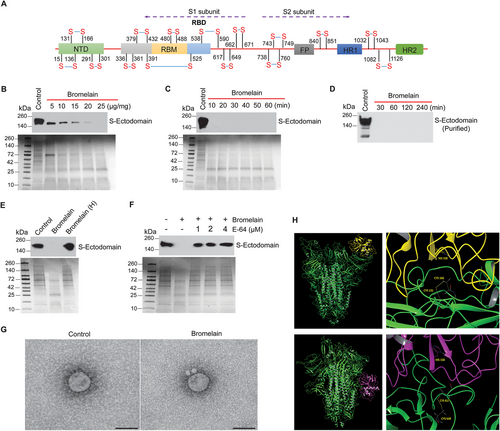
The S-ectodomain has 30 cysteine amino acids with 15 stabilizing disulfide bonds (UniProtKB: P0DTC2) (Fig. 2A). The RBD domain alone has nine cysteine residues, eight of which form four disulfide linkages. Bromelain-induced a dose- and time-dependent cleavage of S-ectodomain in Tni insect cell supernatant (Fig. 2B and C) and purified S-ectodomain (Fig. 2D). Heat inactivation and cysteine proteinase inhibitor (E-64) treatment inhibited bromelain mediated digestion of S-ectodomain (Fig. 2E and F). Further, SARS-CoV-2 with bromelain treatment showed the loss of Spike protein on the viral surface (Fig. 2G). Our docking studies between homotrimeric (RBD containing chain A) S protein and stem bromelain revealed that bromelain cleaves the S-protein equally likely at the 131–166 (50.4%) and 617–649 (49.6%) disulfide bonds (Fig. 2H). Though the catalytic site is not directly engaged, protein–protein docking places the enzyme in close proximity to these bonds. These results demonstrate that bromelain's cysteine protease activity is responsible for the cleavage of host cells’ ACE-2 and SARS-CoV-2 S-protein.
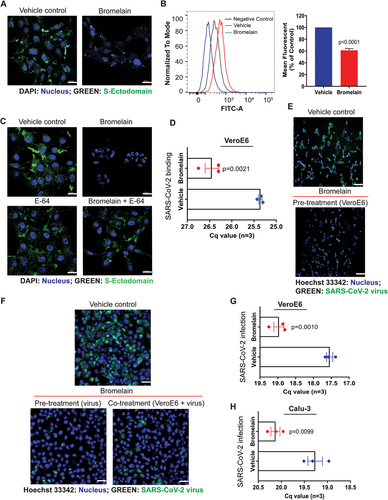
Since bromelain digested ACE-2 and S-ectodomain, we investigated the effect of bromelain on the interactions of S-ectodomain and SARS-CoV-2 with VeroE6 cells. Bromelain significantly reduced the binding of S-protein to VeroE6 cells (Fig. 3A and B) and was further confirmed by cysteine protease inhibitor (E-64) treatment (Fig. 3C). Interestingly, bromelain pre-treatment significantly decreased SARS-CoV-2 viral binding in VeroE6 cells (P = .0021) (Fig. 3D). Most importantly, VeroE6 cells or SARS-CoV-2 or both with bromelain reduces the viral infection (Fig. 3E and F). Additionally, we found significantly reduced SARS-CoV-2 viral RNA copies in bromelain-treated VeroE6 (P = .0010) and Calu-3 (P = .0099) cells (Fig. 3G and H, respectively). Collectively, these results suggest that bromelain could inhibit SARS-CoV-2 binding and infection in VeroE6 and Calu-3 cells. Studies have demonstrated that SARS-CoV-2 S-protein has high homology among other coronaviruses (76% identity with SARS-CoV) with conserved cysteine amino acids (UniProtKB: P59594). This indicates that bromelain may be used as a broad antiviral agent against SARS-CoV-2 and other related family members.
In conclusion, the currently used drugs against SARS-CoV-2 have potential side effects. Vaccine trials have started against COVID-19, but the host immune response against SARS-CoV-2 is not fully understood. It differs between individuals, and also re-infection of individuals with SARS-CoV-2. For the first time, our results demonstrate that bromelain can inhibit SARS-CoV-2 infection via targeting ACE-2, TMPRSS2, and SARS-CoV-2 S-protein. Also, thrombosis development is a significant risk factor of multiorgan failure and death in COVID-19 patients.10 Since bromelain inhibits SARS-CoV-2 infection, and its profound fibrinolytic activity 6 suggests that bromelain or bromelain-rich pineapple could be used as an antiviral against SARS-CoV-2 and future outbreaks of other coronaviruses.
ACKNOWLEDGMENTS
We thank Dr. Kenneth Cowan, Dr. Dalia ElGamal, and Melody J. Morwitzer (UNMC, USA) for their strong support and help. We also thank Elizabeth Coyle (FDA, USA) and Silvie Muschter (Robert Koch Institute, Germany) for their technical assistance. The authors on this manuscript are supported in part by the National Cancer Institute (NIH) grant (R01CA208108), and by the Fred and Pamela Buffett Cancer Center start-up funds (to P.R), and the Fred and Pamela Buffett NCI Cancer Center Support Grant (P30CA036727) (to G.E.O.B.).
CONFLICT OF INTEREST
Satish Sagar and Prakash Radhakrishnan have ownership interest (including patents) in a pending patent.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The institutional review board (IRB) of the University of Nebraska Medical Center approved clinical samples used in this study.
AUTHOR CONTRIBUTIONS
Prakash Radhakrishnan conceived the idea; Surender Khurana, St. Patrick M. Reid, Gloria E. O. Borgstahl, and Prakash Radhakrishnan designed research; Satish Sagar, Ashok Kumar Rathinavel, William E. Lutz, Lucas R. Struble, St. Patrick M. Reid, Tobias Hoffmann, and Mara J. Broadhurst performed research; Nitish Kumar Mishra, Chittibabu Guda, Nicholas Y. Palermo, and Kenneth W. Bayles contributed new reagents/analytic tools; Satish Sagar, Ashok Kumar Rathinavel, Andy T. Schnaubelt, Gloria E. O. Borgstahl, St. Patrick M. Reid, and Prakash Radhakrishnan analyzed data; Gloria E. O. Borgstahl, and Prakash Radhakrishnan wrote the paper.
DATA AVAILABILITY STATEMENT
Materials are available upon a reasonable request from the corresponding author.




