Positive epigenetic regulation loop between AR and NSUN2 promotes prostate cancer progression
Wenkai Zhu, Fangning Wan and Wenhao Xu contributed equally to this study.
Abstract
Background
Prostate cancer (PCa) is a major type of cancer in man worldwide. Androgen deprivation therapy (ADT) and the next-generation androgen receptor (AR) pathway inhibitors have acquired great success in treating PCa. However, patients treated with ADT or AR targeted therapy are inevitably developing into castration-resistant prostate cancer (CRPC) or becoming drug resistance. The role of mRNA 5-methylcytosine (m5C) modification in cancers is largely unknown. This study aimed to explore the role of the m5C methyltransferase NSUN2 in Prostate cancer (PCa).
Methods
The expression of NSUN2 and its clinicopathological impact were evaluated in PCa cohorts. The effect of NSUN2 on the biological characteristics of PCa cells was investigated on the basis of gain-offunction and loss-of-function analyses. Subcutaneous models further uncovered the role of NSUN2 in tumor growth. Epi-transcriptome assays with RNA bisulfite sequencing (RNA-BisSeq) analysis and in vitro enzyme reaction assays were performed to validate the targeted effect of NSUN2 on AR. AR-binding sites in the NSUN2 promoter were investigated by ChIP and luciferase assays to uncover the interplay between NSUN2 and AR signaling. RIP-qPCR and EMSA methods were performed to confirm that YBX1 binds to AR m5C sites.
Results
NSUN2 is highly expressed in PCa and predicts poor outcome. NSUN2 plays roles as a PCa oncogene both in vitro and in vivo. Depletion of NSUN2 results in decreased expression and activities of AR, including AR-V7. Mechanistically, NSUN2 posttranscriptionally stabilized AR by cluster m5C modification in a m5CYBX1-dependent manner. Strikingly, treatment with enzalutamide, an effective AR inhibitor, reduces NSUN2 expression and decreases the m5C modification level in prostate cancer cells. Finally, we found that AR transcriptionally regulates NSUN2.
Conclusion
NSUN2 stabilizes AR mRNA through cluster 5-methylcytosine modification and activates a positive feedback loop to promote prostate cancer.
1 INTRODUCTION
Prostate cancer (PCa) is the most common male malignancy in the West, and its incidence is rapidly increasing in Asia.1, 2 The most important therapeutic target in PCa is the androgen receptor (AR).3 Androgen deprivation therapy (ADT) has long been the cornerstone of PCa treatment, especially metastatic PCa treatment.4, 5 However, metastatic PCa treated with ADT inevitably develops castration-resistant prostate cancer (CRPC).6, 7 Since 2012, the FDA has approved several new AR inhibitors, such as abiraterone, enzalutamide, and apalutamide, to treat CRPC.5, 8-10 These drugs, prolonged the overall survival of PCa patients. However, AR variants such as AR splice variant 7 (AR-V7) and AR-V9 can stimulate AR inhibitor resistance, as they can induce self-activation without androgen binding.11, 12 Therefore, it is really urgent to uncover the resistance mechanisms and develop novel therapeutics.
The mechanisms behind CRPC can be complex. It occurs primarily through AR signalling reactivation in a ligand-dependent or ligand-independent manner.13 Resistance can occur for many reasons. It can be intrinsic, such as a mutation of TP53, or acquired, such as AR amplification or mutations.14 AR amplification or mutations are the most common recurrent somatic gene alterations in mCRPC, accounting for approximately 62.7% in a biopsy study.15 For these reasons, AR is still an important therapeutic target in CRPC treatment.
RNA 5-Methylcytosine (m5C) modifications were first discovered in abundant and stable tRNAs and rRNAs.16 Recently, the mRNA m5C modification has been identified to regulate mRNA metabolism and translocation.17, 18 It has been shown that mRNA m5C modification could impact various biological events, such as embryonic development, myelopoiesis19 and tumorigenesis.17 There are nine subfamilies of cytosine-5 methyltransferases (RCMTs). However, only three subfamilies (RCMT2, RCMT7 and RCMT8) are found in eukaryotic species. NSUN2 and NSUN6 are the two known m5C methyltransferases (‘writers’) of mRNA to date.18, 20, 21 YBX1 and ALYREF, discovered by Yang, are the only known m5C 'readers.'17, 18 NSUN2 has been reported to be involved in promoting the progression of some tumours. It can modify the mRNA of HDGF to promote the progression of bladder cancer.17 In gastric cancer, NSUN2 represses p57Kip2 in an m5C-dependent manner.22 NSUN2 is important for cervical cancer progression.23 Autotaxin mRNA can be methylated by NSUN2 and promote tumour migration and invasion.24 As a new epigenetic regulation mechanism in tumour biology, whether or how NSUN2 plays roles in PCa, even in CRPC and drug resistance remains largely unknown. YBX1 acts as a reader to promote many kinds of cancers that have been reported. YBX1 can form a positive feedback loop with lncRNA to activate the FOXA1 transcription network.25 FOXA1 can interact with AR and help shape the AR signalling.26 These indicate that NSUN2 and YBX1 may act in an axis to influence the AR signalling. The aberrant AR signalling contributes to PCa progression and even its formation.
2 MATERIALS AND METHODS
2.1 Cell lines, antibodies, chemical inhibitors
The human PCa cell lines C4-2, LNCaP, and 22RV1 were used. Details of the antibodies are in Supporting Information. Actinomycin D was purchased from Abcam, Shanghai. Enzalutamide (MDV3100) was purchased from Selleck, Shanghai.
2.2 Cell culture, siRNA and plasmid transfection, lentivirus generation and infection
C4-2, LNCaP, 22RV1 were maintained in RPMI 1640 basic medium (Gibco, C11875500BT) with 10% FBS (Gibco, A3160802).
NSUN2 siRNAs were synthesized by Biosun Company.
Lipofectamine 3000 (Invitrogen) was used for plasmid and siRNA transfection following the manufacturer's protocols.
2.3 RNA m5C quantification by LC/MS/MS
mRNA from C4-2 cells was purified using the Dynabeads™ mRNA Purification Kit. The purified mRNA is digested into single nucleotides and quantitatively detected by LC/MS/MS by Shanghai Biotree Co., Ltd. The content of m5C and cytosine was calibrated by the standard curve generated by the standards (purchased from APExBIO). Following the standard protocol, add 200-300 ng of mRNA to 25 µl of dilution buffer, digest with nuclease P1 (1 U) for 2 h at 42°C, followed by alkaline phosphatase (1 U) and NH4HCO3 (1 M, 3 µl) treated at 37°C incubate for 2 h, and finally filter through a filter for mass spectrometry analysis.
2.4 Dot blot assay
Samples were spotted onto Amersham HybondTM-N+ membranes (GE Healthcare) and cross-linked with UV, then washed with wash buffer for 7 min and blocked for 1 h at room temperature, with anti-m5C antibody (Abcam, ab214727, 1:1000) at 4°C. Then incubated with secondary antibody (Proteintech, SA00001-2) for 1 h and exposed with LAS 4000 mini for 30 seconds.
2.5 Immunocytochemical staining and cell imaging
Cells were fixed with methanol for 15 min and then permeabilized with 0.1% Triton X-100 on ice for 15 min. This was followed by blocking with 10% FBS for 1 h, followed by incubation with primary antibody overnight at 4°C, followed by incubation with secondary antibody for 1 h, and finally mounting with DAPI-containing mounting medium (P36931, Invitrogen). The fluorescence intensity was quantified by ImageJ.
2.6 Secreted PSA quantification
Cells were pre-treated for 72 h. Then, add 50 µl of supernatant to the chip (a rapid quantitative immunoassay analyser) (FREND™ System, NanoEnTek Inc. Korea) and wait for 5 min. Finally, the chip was put into the detection instrument (FREND™ System, NanoEnTek Inc. Korea).
2.7 RNA probe synthesis
The sequence probes were designed in our laboratory. These probes were synthesized by a standard RNA synthesis kit (NEB, E2050) following the manufacturer's protocols.
2.8 RNA pull-down assay
Cell lysates were prepared with RIPA cell lysis buffer. Briefly, mix 50 µl streptavidin magnetic beads with 50 pmol 3′-biotin-labelled RNA probes and incubated for 30 min with agitation. After incubation, add 100 mg protein lysate and incubated for 60 min at 4°C with agitation. The probe sequences were as follows:
probe-mut: AugAugAugAagaagAagAagAagAagaagaagaagaagaagaagaagaagaagaagaagaagaagaagaag
probe-normal: CugCugCugCagcagCagCagCagCagcagcagcagcagcagcagcagcagcagcagcagcagcagcagcag
2.9 Biochemistry assay for RNA m5C transferase reaction in vitro
0.15 nmol probe and 0.15 nmol NSUN2 protein were added into a buffer contain 0.8 mM SAM, 1.5 mM MgCl2, 80 mM KCl, 0.2 U µl−1 RNasin, 0.2 U µl−1 DNasin, 4% glycerol, 10 mM DTT and 15 mM HEPES (pH 7.9) incubated at 16°C overnight.
2.10 Bisulphite conversion
Purified mRNA (500 ng) was converted with the EZ RNA Methylation Kit (Zymo Research).
2.11 RNA-seq
2.11.1 Library preparation and deep sequencing
We collected samples from Fudan University Shanghai Cancer Center (FUSCC). The tissues were put into a homogenization tube, grinding beads and 1 ml of TRIzol (Invitrogen) were added. After grinding, it was centrifuged at 12 000 rpm/min for 15 min, and the supernatant was collected. RNA was purified by TRIzol (Invitrogen). All RNA libraries were constructed with RNA Library Prep Kit for Illumina (E7760), and ribosomal RNA (rRNA) was deleted with the NEBNext rRNA Depletion Kit (E7405). Then, we sent the libraries to a sequencing facility for Illumina 6G sequencing. The raw read qualities were evaluated by FastQC.
2.11.2 RNA-Seq data processing
Raw reads were filtered using Trimmomatic to remove low-quality bases and adaptor sequences and then aligned to the GRCh38 human genome with GENCODE v29 gene annotation using STAR. Gene expression levels were calculated with FPKM values by normalizing gene counts from feature counts. All statistical analyses were performed in R.
2.12 PCR and Sanger sequencing
Five hundred nanograms of in vitro m5C transferase reaction probes, nonreaction probes and negative control R-Luc were converted (see above). Then, the target sequences of the m5C transferase reaction probes, nonreaction probes and R-Luc were amplified by PCR and subjected to Sanger sequencing.
2.13 RIP
The EZ-Magna RIP kit (NO.17-701) was used. Briefly, cells were lysed on ice with RIP lysis buffer. Then, 50 µl protein A/G magnetic beads, 100 µl cell lysis and 900 µl RIP buffer were added to a clean microtube and rotated at 4°C for 3 h. Ten microliters of cell lysate was kept as input and stored at −80°C. Finally, the RNA was purified, collected and converted to cDNA for qPCR.
2.14 ChIP
The ChIP assay was performed with EZ-Magna ChIP G (Millipore, 17–409). Briefly, the cell lysates were prepared with cell lysis buffer and nuclear lysis buffer with protease inhibitor cocktail II. Then, the cell lysates were ultrasonicated to break the genomic DNA into 200 -1000 bp pieces. After that, the treated cell lysates, antibodies and protein G magnetic beads were mixed and incubated at 4°C with rotation. Finally, the DNA fragments were purified and collected for qPCR and sequencing.
2.15 mRNA stability assay
Each sample was harvested at 0, 3, 6, and 7 h after treatment with actinomycin D (2 µM). Total RNA was isolated and converted to cDNA for RT-qPCR analysis.
2.16 Luciferase reporter assay
The modification site sequence of AR was inserted downstream of the promoter of the pGL3-promoter luciferase vector (Vigenebio, Maryland, USA). C4-2 cells were co-transfected with a mixture of 2000 ng pGL3-promoter-AR-Luciferase reporter vector and 1000 ng pRL-TK vector. The relative luciferase activity was measured with a dual luciferase reporter assay system (Promega, Madison, WI).
2.17 EMSA
Biotin-labelled RNA oligonucleotides were produced according to the above protocol. Recombinant human YBX1 proteins were purchased from Abcam (ab187443). The YBX1 proteins were mixed with probes in a binding buffer for 30 min at room temperature. The product was separated with 6% native PAGE gel and transferred to a nylon membrane. Membranes were then UV cross-linked, blocked for 15 min, and incubated with HRP-linked streptavidin (1:300) for 15 min. Add chemiluminescent substrate and incubate for 1-3 min to expose with LAS 4 000 mini for 1-5 min. The probe sequences were as follows: 5′-cuuuccagaaucug……cugcugcugcagcagcagcagcagcagcagcagcagcagcagcagcagcagcagcagcagcagcagcagcagcagcaagagacuagccccaggcagcagcagcagcagcagggugaggaugg……gccagcaaggggcugcc-3′.
2.18 In vivo xenograft assay
This study was approved by Fudan University (2020 FUSCC JS-243). All treatments were administered according to the guidelines of the Institutional Animal Care and Use Committee.
A total of 107 C4-2 (sh-NSUN2-2, OE-NSUN2 or control) cells were mixed with Matrigel and injected into the backside of 6-week-old male BALB/cA nude mice. The animals were checked every 5 days. The tumours were weighed after the mice were sacrificed.
2.19 Patients collection
A total of 12 452 cancer patients with available level 3 RNA-seq data were recruited from the TCGA database in the website: http://xena.ucsc.edu. Clinical data and gene expression profiles of 497 PCa tissues and 52 normal tissues were obtained. Besides, 88 high-risk PCa patients enrolled from FUSCC, with pathology and electronic medical records provided. Samples of PCa and normal prostate tissues were collected during surgery or biopsy, then processed and stored at the FUSCC tissue bank.
2.20 Immunoblotting
Primary antibodies against NSUN2, AR, AR-V7, YBX1 and β-actin were used for the immunoblot assay in this study. Detailed information is listed in Supporting Information.
2.21 Statistical analysis
Statistical analysis was performed with SPSS 22.0 software (SPSS, Chicago, Illinois) and R. Continuous variables were compared with a t-test, while categorical variables were compared with a χ2 test. All significance tests were two-sided, and differences with a p-value < .05 were considered significant. The cut-off value was defined via median value or using the 'survminer' R package.
3 RESULTS
3.1 NSUN2 upregulation in PCa associated with worse prognosis
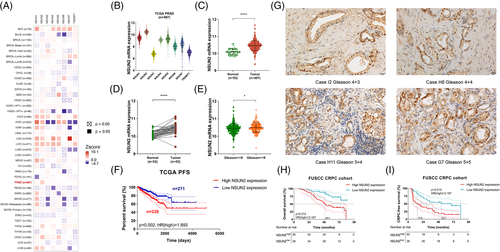
NOP2/Sun RNA methyltransferase gene families and TRDMT1 are known human cytosine-5-methyltransferases responsible for the methylation of RNA carbon 5 in cytidine (m5C, 5-methylcytidine).27-29 In the Cancer Genome Atlas (TCGA) pan-cancer result, NSUN2 was the only poor outcome predictor of overall survival (OS) in PCa (Figure 1A). Also, in the TCGA prostate adenocarcinoma (PRAD) cohort, NSUN2 was the most abundant gene among the cytosine-5-methyltransferases in PCa (Figure 1B). The clinicopathological parameters were shown in Table S1. NSUN2 mRNA expression levels were significantly higher in 497 tumour samples than in 52 normal tissues (p < .01, Figure 1C). In 52 paired PCa and adjacent normal tissues, NSUN2 mRNA expression levels were markedly elevated in tumour tissues (p < .01, Figure 1D). NSUN2 mRNA expression significantly increased in patients with high Gleason scores (p < .05, Gleason > = 8, Figure 1E). High NSUN2 expression was associated with shortened PFS in 497 TCGA PCa patients (p = .002, HR = 1.893, Figure 1F) and shortened OS in 492 TCGA PCa patients (p = .048, HR = 4.2, Figure S1A). In contrast, NSUN6 showed no outcome predictive values in either OS (p = .63, HR = 0.74, Figure S1B) or DFS (p = .47, HR = 1.2, Figure S1C) in the same cohort. We performed immunohistochemistry (IHC) staining of a cohort of 88 high-risk PCa patients obtained from the Fudan University Shanghai Cancer Center (FUSCC). The clinicopathological parameters were shown in Table S2. Representative images were shown in Figure 1G. High NSUN2 expression was associated with poor OS (p = .012, HR = 3.187, Figure 1H) and shorter CRPC-free survival (p < .001, HR = 2.467, Figure 1I). These results demonstrated that the upregulation of NSUN2 is a frequent oncogenic event in human PCa patients.
3.2 NSUN2 regulates the proliferation, invasion and migration of PCa cells in vitro and in vivo
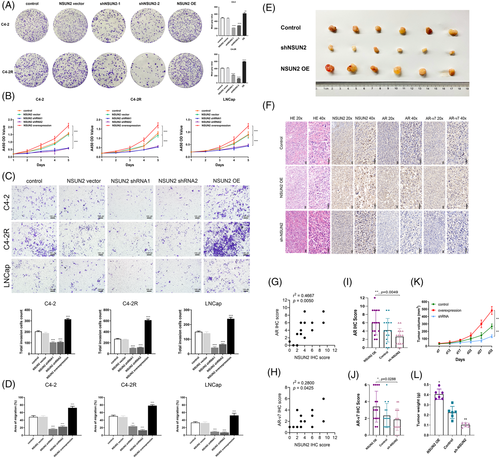
To investigate whether NSUN2 could influence the progression of PCa and be a potential therapeutic target, a series of experiments were conducted. LNCaP (androgen-sensitive PCa cell), C4-2 (castration-resistant PCa cell) and C4-2R (enzalutamide resistant PCa cell)7 cells were transfected with shRNAs targeting NSUN2 or lentiviral overexpression (OE) vectors. Colony formation assays revealed markedly fewer colonies when NSUN2 knocked down, consistently, more colonies when NSUN2 overexpression (Figure 2A). The CCK-8 assay showed that, in the NSUN2 overexpression group, the cell proliferation ability was significantly increased, while in the NSUN2 knockdown group, it was significantly decreased. (Figure 2B). Transwell assays showed a significantly decreased number of invading C4-2, C4-2R or LNCaP cells in the shRNA groups and an increased number of invaded C4-2, C4-2R or LNCaP cells in the NSUN2 OE group compared with the negative control group (Figure 2C). The wound healing assay showed that, in the NSUN2 OE group, the cell migration ability was significantly improved, while in the NSUN2 knockdown group, it was significantly reduced (Figure 2D, Figure S2). In the NSUN2 OE group, the tumour volume was significantly increased, while in the NSUN2 knockdown group, the tumour volume was significantly decreased (Figure 2E). Haematoxylin-eosin (HE) staining and IHC were implemented to detect the expression levels of NSUN2, AR and AR-V7 using tumour tissues harvested from xenograft model mice (Figure 2F). IHC of tumour tissues from the negative control group showed that the expression of NSUN2 was correlated with the expression of AR (r2 = 0.4667, p = .0050) (Figure 2G) and AR-V7 (r2 = 0.2800, p = .0425) (Figure 2H). The IHC scores of AR (Figure 2I) and AR-V7 (Figure 2J) were markedly decreased in the NSUN2 shRNA group and increased in the NSUN2 OE group, indicating that NSUN2 significantly regulates the expression of AR and AR-V7. Decreased tumour volume and weight were observed in the NSUN2 shRNA group, and increased tumour size was observed in the NSUN2 OE group compared with the control group. Tumour volume (Figure 2K) and weight (Figure 2L) were markedly decreased in the shRNA-treated group and increased in the NSUN2 OE group compared with the negative control group. In conclusion, NSUN2 could influence the proliferation, invasion and migration of PCa cells both in vitro and in vivo. NSUN2 has a significant positive correlation with AR and AR-V7 expression.
3.3 Silencing NSUN2 decreased m5C levels and downregulated AR expression and signalling activity
NSUN2 normally functions as a methyltransferase that catalyses the methylation of cytosine to 5-methylcytosine at RNA. Subcutaneous xenografts IHC results showed that NSUN2 is co-expressed with AR and AR-V7. The immunofluorescence assay in NSUN2 knockdown or overexpression C4-2 cells showed that the m5C fluorescence signal was significantly positively correlated with NSUN2 expression (all p < .05, Figure 3A-C). These results indicated that NSUN2 was responsible for the m5C modification level in C4-2 cells. Consistently the AR mRNA level was decreased when NSUN2 was downregulated by siRNAs (Figure 3D,E). Additionally, silencing NSUN2 led to a decrease in AR protein levels in both C4-2 and LNCaP cells (Figure 3F,G). AR-V7 is encoded by a transcript variant of AR with different splicing at the pre-mRNA level and was reported to influence enzalutamide resistance in PCa.12 We found that the protein level of AR-V7 was downregulated after knockdown of NSUN2, suggesting that NSUN2 may regulate AR at the pre-mRNA level (Figure 3H). We designed primers at each intron of AR pre-mRNA (Figure S3A) and found each intron of AR pre-mRNA expression was decreased while NSUN2 was silenced (Figure S3B–H). Furthermore, we found that the expression level of NSUN2 could affect the tolerance of C4-2 cells to enzalutamide (MDV3100) (Figure 3I). At the same time, the proliferation and migration ability of 22RV1 cells were also affected by the expression level of NSUN2 (Figure S3 I–K). Dihydrotestosterone (DHT) can promote the development of the prostate and accelerate the AR transfer into the nucleus, functioning as a transcription factor. Through CCK-8 (Figure 3J) and clone formation assays (Figure 3K) with NSUN2 knockdown LNCaP cells treated with DHT, we found that DHT can rescue the NSUN2 expression (Figure S3L) and the cell proliferation ability of NSUN2 knockdown cells. Therefore, NSUN2-mediated regulation of PCa progression may be dependent on AR signalling.
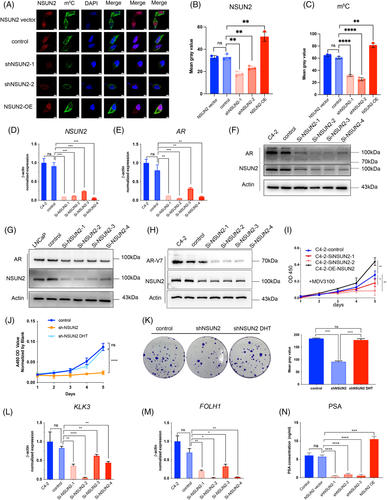
To detect whether the AR signalling activity was also influenced by NSUN2 expression, we detected KLK3 (known as prostate-specific antigen [PSA]) and FOLH1 (known as prostate-specific membrane antigen [PSMA]) at the transcriptional level by quantitative real-time PCR (RT-qPCR). The transcriptional levels of KLK3 and FOLH1 were decreased by siRNA-mediated silencing of NSUN2 (Figure 3L,M). The PSA protein levels in the C4-2 cell supernatant were changed along with NSUN2 expression, as indicated by ELISA (Figure 3N).
3.4 NSUN2-mediated m5C modifications are clustered at the 5′ end of AR mRNA
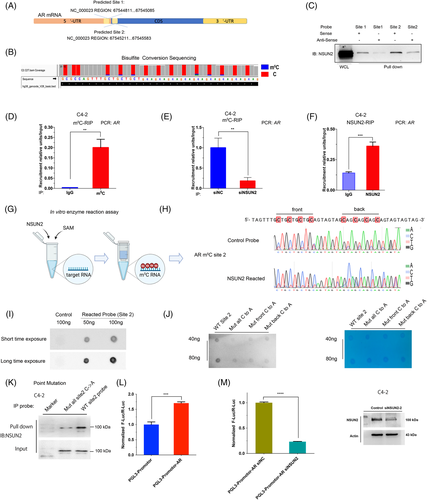
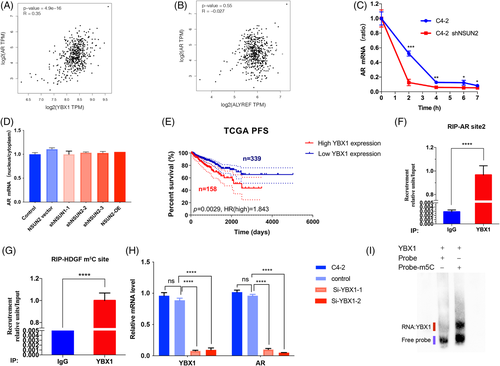
Given that NSUN2 silencing attenuated the expression of AR at both the transcriptional and protein levels, and AR signalling activity, we hypothesized that m5C modification may exist in AR mRNA. We performed RNA bisulphite sequencing (RNA-BisSeq) in C4-2 cells and observed multiple unconverted cytosines near the end of the 5′-UTR and start codon of AR (Figure 4A,B). We called the m5C cluster near the end of the 5′-UTR 'site 1' and the m5C cluster near the start codon 'site 2'. We next performed the pull-down assays with synthesized probes of site 1 and site 2. The results showed that both site 1 and site 2 RNA could bind to the NSUN2 protein in C4-2 cells. Since the site 2 m5C signals in the BisSeq and pulldown assays were stronger than those for site 1, the following experiments were conducted using site 2 (Figure 4C). RNA immunoprecipitation (RIP) assays showed that the m5C antibody could enrich AR mRNA in vitro (Figure 4D), whereas after silencing NSUN2, less AR mRNA was enriched in the m5C-RIP assay (Figure 4E). The NSUN2-RIP assay showed that the NSUN2 antibody group could markedly enrich AR mRNA in C4-2 cells compared with the IgG group (Figure 4F). To test the reliability of the above experiment, we performed the same test with a known HDGF m5C modification site17 (Figure S4A,B). Previous reports discussed only the NSUN2 mononucleotide activity as an mRNA methyltransferase.30 To test whether NSUN2 is able to modify m5C of AR mRNA in clusters, we carried out the in vitro enzyme reaction assays using in vitro-transcribed fragments of AR mRNA, recombinant NSUN2 protein, and S adenosyl-L-methionine (SAM) (Figure 4G). After the enzyme reaction, the probes of site 2 were examined by Sanger sequencing upon bisulphite conversion, and we found multiple C signals for NSUN2-reacted probes that were stronger than those for the control probes (Figure 4H). The dot blot assay was conducted to confirm the reaction assay. The reacted probes could enrich the m5C antibodies, but the untreated control probes could not (Figure 4I). In order to assess that the clustered C sites on AR mRNA are critical for the enzymatic activity of NSUN2, we mutated the first four Cs and the last four Cs individually or all. An in vitro enzyme assay demonstrated that mutation of multiple C sites decreased the m5C level of AR site 2 RNA probes (Figure 4J). Mutated and wild-type site 2 probes were used in the pulldown assay and found that site 2 cytosines were found to be crucial for NSUN2 binding (Figure 4K). Finally, the AR site 2 sequence was inserted into the PGL3-promoter plasmid. The group with the AR site 2 sequence showed stronger luciferase activity than the control group (Figure 4L) and was dependent on the presence of NSUN2 (Figure 4M).
3.5 YBX1 is the reader of AR mRNA m5C modification
To investigate the recognition mechanism of AR mRNA m5C modification, we first analysed the already known m5C 'readers,' YBX1 and ALYREF. As reported, ALYREF participates in mRNA export and YBX1 plays a role in mRNA stability.17, 18 A scatter plot of YBX1 expression and AR expression in the TCGA PRAD cohort (N = 497) indicated that YBX1 expression was positively correlated with AR expression (p = 4.9e-16, Figure 5A). However, ALYREF expression was not correlated with AR expression (p = .55, Figure 5B). Silencing NSUN2 influenced AR mRNA stability (Figure 5C) but did not influence RNA export from the nucleus to the cytoplasm (Figure 5D). Clinically, elevated YBX1 expression correlated with shortened PFS in 497 patients from the TCGA cohort (p = .0029, HR = 1.843, Figure 5E). RIP assays showed that YBX1 could bind AR mRNA (Figure 5F) and HDGF was used as a positive control (Figure 5G). Additionally, silencing YBX1 in C4-2 cells led to decreased AR mRNA (Figure 5H). The luciferase activity was decreased when knockdown YBX1 (Figure S4C). This may not be related to the transcriptional activity of YBX1, because YBX1 could not be a transcription factor of AR (Figure S4D). Furthermore, we used an electrophoretic mobility shift assay (EMSA) to assess the binding abilities of the YBX1 protein with m5C modified or unmodified AR site 2 probes (20 ng). YBX1 could only bind m5C-modified probes (Figure 5I). The pulldown assay came to the same conclusion (Figure S4E).
3.6 AR regulates m5C and NSUN2 in PCa
In Gene Expression Omnibus (GEO) dataset (GDS4120), we found that NSUN2 expression levels in mice LuCaP35 xenografts decreased after 4 weeks of surgical castration31 (Figure 6A). In the GDS3358 dataset, NSUN2 expression levels in androgen-deprived LNCaP decreased from 3 weeks to 5 months and rebounded in 11 months32 (Figure 6B). Notably, AR expression was positively correlated with the NSUN2 level in the TCGA PRAD dataset (Figure 6C). In FUSCC PCa samples, NSUN2 expression was greatly altered during the period of ADT. NSUN2 expression decreased in an initial ADT for 3 months, whereas abiraterone-resistant PCa samples showed markedly higher expression of NSUN2 (Figure 6D). These analyses together suggest that AR may play an important role in regulating NSUN2, thereby the formation of m5C RNA. MDV3100 (enzalutamide) is a newly developed AR antagonist.10 We used enzalutamide to block AR activity in C4-2 cells for 72 h and found that the m5C levels were dramatically decreased in a dot blot assay (Figure 6E). Consistently, RNA m5C quantification by LC/MS/MS showed similar results in C4-2 cells (Figure 6F, Figure S4F,G). Interestingly, the expression of NSUN2 and AR decreased in C4-2 cells upon enzalutamide treatment (Figure 6G,H) and apalutamide treatment (Figure S5A,B). We next conducted a fluorescence immunocytochemical staining assay and performed a live cell imaging confocal microscopy analysis. The results showed that the NSUN2 signal was mainly distributed in the nucleus, while the m5C signal was mainly distributed in the cytoplasm of C4-2 cells (Figure 6I). Enzalutamide treatment significantly decreased NSUN2 expression and m5C levels (Figure 6J,K). In xenograft mouse model, shNSUN2 or treatment with apalutamide can inhibit tumour proliferation, and the combination of shNSUN2 and apalutamide is more effective (Figure S5C–E). These results support that AR inhibition downregulates NSUN2 expression and decreases the mRNA m5C level. The combination of shNSUN2 and AR inhibitors may become an effective clinical treatment.
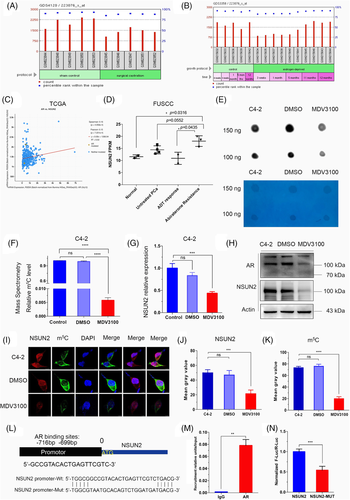
We further investigated whether AR directly regulates NSUN2 transcription. So, we predicted the AR DNA-binding motifs in the NSUN2 promoter sequences using the JASPAR database (http://jaspar.genereg.net).33 The AR binding site was revealed to be a 19-bp sequence, approximately 700 bp upstream of the transcriptional start site (TSS) of NSUN2 (Figure 6L). ChIP-qPCR assays also showed significant enrichment of AR in the NSUN2 promoter region (p < .05, Figure 6M). To further validate the binding site, we constructed a luciferase plasmid with wild-type and mutated AR binding sites in the NSUN2 promoter. AR binding site was verified by dual-luciferase analysis (p < .01, Figure 6N). Apalutamide can decrease luciferase activity (Figure S5F). ChIP-seq for H3K27ac profiling of PCa samples showed that the promoter region of NSUN2 was active in PCa samples 34, 35(Figure S6A). Chromatin immunoprecipitation sequencing (ChIP-seq) profiling of AR genome-wide binding sites demonstrated that the NSUN2 promoter region has significant AR binding signals in both PCa cell lines (Figure S6B) and tumour samples (Figure 6C).
3.7 NSUN2 is positively correlated with AR signalling genes and predicts poor outcomes in PCa
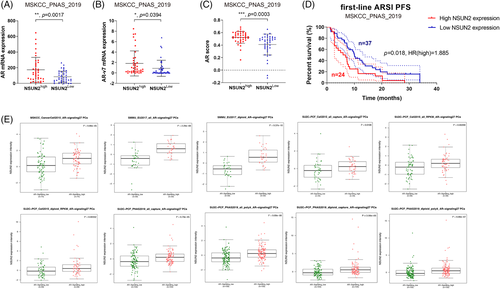
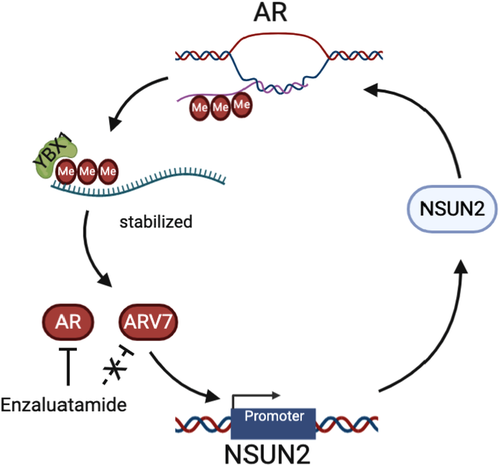
To evaluate the expression patterns and prognostic values of NSUN2 in PCa, external validation cohorts were analysed. Significantly elevated AR (Figure 7A) and AR-V7 (Figure 7B) mRNA expression and AR scores (Figure 7C) were found in the high NSUN2 expression group compared with the low NSUN2 expression group among 91 PCa patients from the Memorial Sloan Kettering Cancer Center (MSKCC) cohort8 (p < .05). Most of these patients received first-line second-generation androgen receptor signalling inhibitor (ARSI) treatment. The progression-free survival (PFS) after ARSI was plotted according to the level of NSUN2 expression. High NSUN2 expression significantly predicted poor PFS for 61 PCa patients from the MSKCC cohort (p = .018, HR = 1.885, Figure 7D). We analysed data for 598 PCa patients from multiple cohorts and found that increased NSUN2 mRNA expression was significantly correlated with elevated AR signalling scores 8, 15, 36, 37 (Figure 7E). In conclusion, NSUN2 can influence the expression of AR and affect the effect of ARSI treatment. On the other hand, AR also had some impact on NSUN2 expression (Figure 8).
4 DISCUSSION
The role of mRNA m5C modification in cancers is largely unknown.17 Previous studies indicated that dynamic m5C modifications are associated with testis development in mice.18, 21 Testis development is dependent on AR expression and the AR signalling pathway. Also, AR is an important therapeutic target in PCa management. In addition, investigators have found that the signal of m5C modifications decreased in enzalutamide-treated C4-2 cells, so we tried to uncover the functions of m5C modifications in PCa. Our data provide evidence that NSUN2 is highly expressed in PCa and is associated with poor prognosis. Mechanistically, NSUN2 post-transcriptionally stabilized AR by cluster m5C modification in an m5C-YBX1-dependent manner. At the same time, AR acts as a transcription factor to regulate the transcription of NSUN2. The positive feedback between NSUN2 and AR provides a possible model to explain PCa progression and the occurrence of CRPC.
RNA modifications like m6A have been reported to regulate diverse cellular functions in many tumours.38 Recently, the mRNA m5C modification has been identified to regulate mRNA metabolism and translocation.17, 18 NSUN2 and NSUN6 are the two known m5C methyltransferases (‘writers’) of mRNA to date.18, 20, 21 However, we found only NSUN2 can be the poor outcome predictor of PCa. NSUN2-related m5C modification sites on mRNA are generally located in the translational start sites, 3′ untranslated regions (UTRs).39, 40 We found the m5C sites were clustered in the 5′ end of AR mRNA and maintains the stability of AR mRNA. Traditional analysis of m5C modification sites used to ignore the cluster C signal for two reasons. First, false positives may occur due to incomplete bisulphite conversion in the GC-rich sequence, which more easily forms secondary structures.41, 42 Second, the validation of the mono m5C site by point mutation is much easier than that of cluster m5C sites. In this study, we applied multiple experiments to illustrate that AR mRNA has a cluster of m5C modifications in the 5′-end regions. We used Sanger sequencing and dot blot methods to illustrate this phenomenon. This study provides novel evidence that m5C modification clusters exist and have functions. Previously, a study reported that NSUN2 stabilizes CDKN2B/p16 mRNA.43 However, p16 is an inhibitor of CDK. It seems that NSUN2 would rather lead to reduce cell proliferation through p16. This contradicts the conclusion in this study that NSUN2 promotes PCa cell proliferation. Then, we found that the expression of NSUN2 was more strongly associated with AR than with p16 in PCa (Figure S7A). When NSUN2 was knocked down in C4-2 cells, the expression level of AR was significantly reduced, while the change of p16 was not so obvious (Figure S7B). Therefore, we hypothesized that the effect of NSUN2 on p16 in PCa is not sufficient to influence NSUN2 to promote PCa cell proliferation through AR. At the same time, we found that high expression of p16 in PCa is associated with poor DFS (Figure S7C,D). So, we predict that NSUN2 could promote PCa cell proliferation, mostly. Also, NSUN2 could methylate many other RNAs like from our data, such as FOXA1 and SMAD3 (Figure S7E–G). These genes could also contribute to the poor prognosis of PCa. Particularly, m5C added by NSUN2 has been detected not only in mRNA but also in rRNA and tRNA.18, 39 Recently, researchers have reported that NSUN3 is participating in the mitochondrial tRNA modifications and shaped metabolic plasticity in metastasis of oral cancer.44 It deserves further researches whether NSUN2 could influence the metabolism and oxidative stress through mitochondrial tRNAs.
Historically, ADT with or without first-generation AR inhibitors such as flutamide and bicalutamide has been the standard of care for metastatic castration-sensitive PCa.45 In this stage, the majority of patients have an initial response to ADT. However, most men with metastases develop CRPC in a median time frame of approximately one year.4, 46 We found NSUN2 expression levels followed the same trends in the previous studies31, 32 and our cohort. The success of the LATITUDE and ARCHES studies supports the hypothesis that more effective inhibition of AR signalling as the initial systemic therapy in patients with castration-sensitive PCa leads to improved outcomes.5, 9, 47 However, metastatic PCa is still considered a lethal disease, and aberrations of the AR, steroidogenic parallel pathways, and neuroendocrine differentiation are the main reasons for resistance to next-generation ADT.11, 12
AR-V7 leads to ligand-independent constitutive activation that is not inhibited by antiandrogen therapies. Among men with metastatic CRPC, the circulating tumour cell (CTC) AR-V7+ rate was 8%.48 Previous reports suggested that CTC AR-V7 detection is a poor prognostic indicator for the clinical efficacy of secondary hormone therapies, including abiraterone, enzalutamide or galeterone.48, 49 Outcome-associated AR perturbations include AR variants and AR gene alterations.3 These aberrant AR products escape the regulation of any clinically available ARSIs. Our results suggest that AR pre-mRNA can be modified by NSUN2 and maintained stable with the recognition and help of YBX1. Our study indicates NSUN2- m5C-YBX1 axis could regulate ARV7 expression levels as well. Disruption of the NSUN2-m5C-YBX1 axis may interfere with the progression of CRPC due to AR variants and AR genomic alterations. This showed a novel mechanism that regulates all AR perturbations and may be therapeutic in AR-related CRPC. However, there are currently no known specific inhibitors or antibodies for NSUN2 that can be used for clinical research. At present, there is a technology called PROTAC that uses the cell's own ubiquitination degradation mechanism to target and degrade specific proteins, which may be used for the inhibition of NSUN2, but further research is needed. As an important part of this regulatory axis, YBX1 may be an important target for PCa therapy. YBX1 has been reported to promote the progression of many kinds of cancers. Endogenous tRNA-derived fragment displacement by YBX1 can suppress breast cancer progression.50 YBX1 acts as a reader to promote oestrogen resistance in progressive breast cancer cells, and can be inhibited by YBX1 phosphorylation inhibitor TAS0612 (a multi-kinase inhibitor) and everolimus (a rapamycin complex 1 inhibitor).51 LncRNA can form a positive feedback loop with YBX1 to activate the FOXA1 transcription network in cancer.25 More importantly, FOXA1 is also an important transcription factor in PCa and is closely involved in the regulatory network of AR. Our data also shows that YBX1 can bind and stable the AR pre-mRNAs which have been modified by m5C. Maybe, YBX1 is a good therapeutic target for PCa and even mCRPC.
In conclusion, we found that m5C modifications exist on AR mRNA. NSUN2 can methylate AR pre-mRNA and influence AR-V7 expression, and YBX1 can bind and stable the m5C modified AR mRNA. AR can promote NSUN2 transcription. The NSUN2 and AR feedback loop leads to a poor outcome in PCa due to ARSI resistance. These findings suggest NSUN2 and YBX1 as new targets for the treatment of PCa.
ACKNOWLEDGEMENTS
We thank Professor Gonghong Wei for kindly advices for this study. We thank Jun Wang providing C4-2R cell lines. National Natural Science Foundation of China to Dingwei Ye (81872099), Natural Fund Project of Shanghai Science and Technology Commission to Dingwei Ye (1851110800), Shanghai Municipal Health Bureau (2020CXJQ03) to Dingwei Ye
CONFLICT OF INTEREST
The authors declare no conflict of interest.




