Single-cell RNA landscape of cell fate decision of renal proximal tubular epithelial cells and immune-microenvironment in kidney fibrosis
Minyan Zhu, Zhenhua Zhang, Zhejun Chen and Yao Xu have contributed equally to this work and share first authorship.
This study found that proximal tubular epithelial cells (PTECs), the main population of cells in the kidneys, exhibited transcriptional heterogeneity during interstitial fibrosis and tubular atrophy (IFTA), which contributes to the complex metabolic and immuno-microenvironment. The activated transcription factor (TF) NR1H4 regulated the differentiation and maladaptive repair of PTECs by targeting the phenylalanine hydroxylase (PAH) gene.
IFTA is a shared pathological change that determines the transition and progression of chronic kidney disease (CKD).1 The mechanism underlying the maladaptive repair of PTECs and their crosstalk with the immuno-microenvironment remain unclear. To better understand the transcriptional dynamics and cell composition in IFTA, we characterized the molecular profiles of 21 466 cells from kidney biopsy samples of one donor, two mild IFTA (one membranous nephropathy and one lupus nephritis) and two moderate-to-severe IFTA (one IgA nephropathy and one chronic tubulointerstitial nephritis) through scRNA-sequencing. Clinical information, annotated cell clusters and methods can be found in Supplementary Methods, Tables S1 and S2, Figures 1A,B and S1–S8.
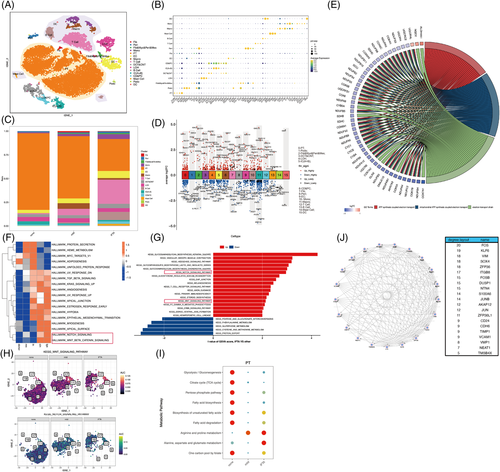
We found that samples with IFTA had fewer PTECs, with more immune cells than that present in other samples (Figures 1C and S4). Enrichment analysis based on differentially expressed genes (DEGs) of PTECs in IFTA versus the others indicated significantly lower energy metabolism and higher mRNA catabolic processes, ribosome-related pathways, cell apoptosis and biosynthesis of amino acids (Figures 1D-E1 and S9). A GSVA and AUCell analysis of all genes between IFTA and the others demonstrated that the amount and activity of hedgehog, notch and WNT pathways (related to cell development in renal fibrosis2) were significantly elevated (Figures 1F–H and S9). Lower glucose and lipid metabolism and higher arginine and glutamate-related metabolism were further found in the PTECs of the IFTA group by the scMetabolism and ssGSEA analysis (Figures 1I and S10a–e), indicating a possible relationship between protein metabolism and the cell differentiation of PTECs. In contrast, TCA cycle was elevated in the mast cells of the mild group, whereas fatty acid degradation and one-carbon metabolism elevated in the T cells of the IFTA group, and pentose phosphate pathway elevated in the B cells of the IFTA group, suggesting probable changes in cell states and the metabolic microenvironment. FOS (regulating TGF-β-mediated signalling3), KLF6 (mediating branched-chain amino acid catabolism loss in PTECs4), VIM and SOX4 (related to the differentiation of PTECs5) were the top hub genes related to IFTA grades found by WGCNA (Figures 1J and S11).
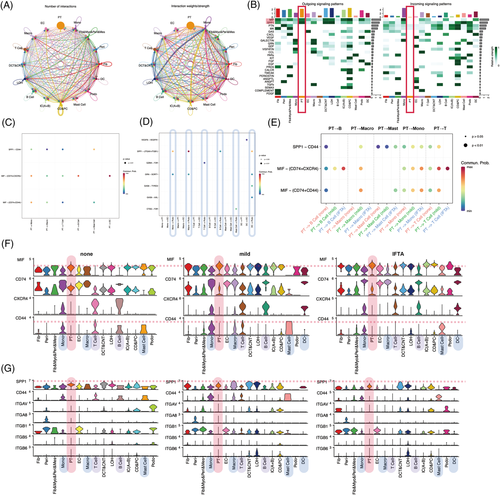
To determine whether heterogeneous PTECs contribute to different immuno-microenvironments during the progression of IFTA besides the probable effects of metabolism, cell crosstalks were calculated by CellChat (Figures 2 and S12–16). PTECs interact with immune cells mainly by the MIF and SPP1 pathways, and the strongest interactions were with monocytes in all samples (Figures 2A,B and S12e). MIF-(CD74 + CXCR4) took most account of MIF pathway from PTECs which were the strongest to dendritic cells (DCs) (Figures 2C and S13f). PTECs received the main signalling of PTN and MK from (myo)fibroblast-like cells instead of immune cells (Figures 2D and S12e, S14). As IFTA progressed, the communication of PTECs with immune cells increased by activating CXCR4 in monocytes, macrophages, T cells, B cells and DC cells, and CD44 in monocytes, macrophages, T cells and mast cells with MIF (Figure 2E–G). The activation in monocytes and macrophages was the most prominent among all immune cells (Figures S15d and 16d). The hyperactivation of CXCR4 and CD44 receptors in immune cells is proinflammatory and promotes kidney injury.6
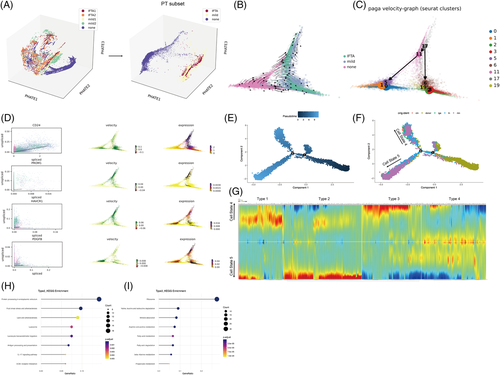
PHATE 3D also visualized that PTECs were the main cell cluster that underwent cell differentiation as IFTA progressed (Figures 3A and S17a). Two Seurat clusters (17, 11) of high capacity were further found to differentiate into normal cells in the none group or clusters (2, 3, 6, 19) with the highest expression of injury molecule (HAVCR1 and PDGFB) (Figures 3B–D and S17b–d) through the RNA velocity and PAGA algorithm. The pseudotime trajectory analysis was next performed using Monocle2 to determine the DEGs and pathways related (Figures 3 and S18). PTECs were differentiated into two different branches (states), where Seurat clusters 2 and 3 (most of which were from the mild or IFTA group) were the downstream of differentiation (Figures 3E,F and S18b). The cells of Seurat clusters 2 and 3 (belonging to cell state 5) showed high protein processing in the ER, fluid shear stress and proinflammatory pathways but low fatty acid degradation (Figure 3G–I).
To determine the potential TFs regulating the transcriptional heterogeneity and differentiation of PTECs, SCENIC analysis was conducted (Figures 4 and S19–28). HNF4A (579 g), NR1H4 extended (191 g), NR2F1 extended (295 g), HNF1A (358 g) and POLE4 extended (431 g) were the top five specific regulons (TF-target gene) in PTECs, among which NR1H4 extended (191 g) and HNF4A (579 g) (that regulates adaptive repair of PTECs7) were the most active, as determined by the average and binary algorithm (Figures 4C and S22). Tubular NR1H4 expression was negatively correlated with eGFR (r = −.537, p = .004), whereas HNF4A positively, through analysing public RNA-seq data8, 9 (Figure S23a,b). The PTECs highly expressing NR1H4 also secreted more proinflammatory CCL15 and C3 to immune cells and showed lower glucose and fatty acid metabolism than the PTECs lowly expressing NR1H4 (Figures 4D and S26, S27). A Venn diagram and motif enrichment analysis showed that NR1H4-PAH (monocle2 type3 gene) had a binding motif of high confidence (details in Table S6), also predicted by the JASPAR database (Figure S24e). NR1H4 levels were high, and PAH levels were low in mice with folic acid-induced nephropathy (FAN) and H2O2-stimulated HK-2 cells (Figures 4F and S28a,b). Mice in which NR1H4 was knocked out or administered the NR1H4 inhibitor Z-guggulsterone or Gly-β-MCA demonstrated less IFTA and apoptosis of PTECs with partially recovered PAH expression (Figures 4G–K and Figure S28c) in FAN. These results were similar to our previous findings on the ischaemia-reperfusion model.10 The HK-2 cells in which PAH was knocked down underwent more severe apoptosis under H2O2 modelling (Figure S28e).
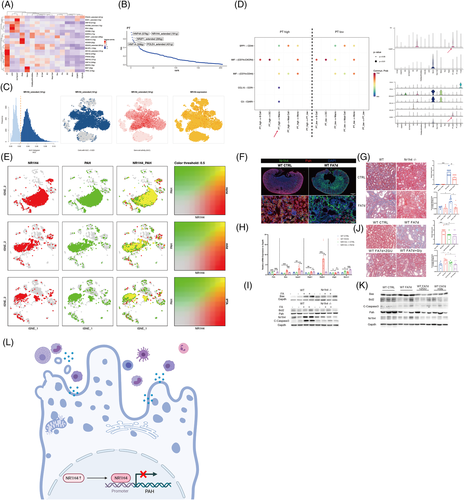
In summary, we found that heterogeneous PTECs contribute to different metabolic and immuno-microenvironments during IFTA. NR1H4 can regulate the maladaptive repair of PTECs through PAH (Figure 4L). These findings provide information for further therapeutic studies on CKD.
ACKNOWLEDGEMENTS
We acknowledge our thanks to the suggestion from Professor Luonan Chen of Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences in searching clinical importance of the transcriptional factors. We also want to acknowledge our thanks to the Innovative Research Team of High-Level Local Universities in Shanghai, and sequencing core of Shanghai Institute of Immunology.
FUNDING INFORMATION
This study was supported by the National Natural Science Foundation of China (81970574, 82170685), as well as a grant (ZXYXZ-201904) from the Science and Technology Commission of Shanghai Municipality, China, and a grant from Shanghai Municipal Health Commission Three-Year Action Plan for Traditional Chinese Medicine (2021-2023). The study was also supported by the Open Project Program Foundation of Key Laboratory of Liver and Kidney Diseases (Shanghai University of Traditional Chinese Medicine), Ministry of Education (GS2022-01).
CONFLICTS OF INTEREST
The authors declare no competing interests.




