High expression of CD38 and MHC class II on CD8+ T cells during severe influenza disease reflects bystander activation and trogocytosis
Abstract
Objectives
Although co-expression of CD38 and HLA-DR reflects T-cell activation during viral infections, high and prolonged CD38+HLA-DR+ expression is associated with severe disease. To date, the mechanism underpinning expression of CD38+HLA-DR+ is poorly understood.
Methods
We used mouse models of influenza A/H9N2, A/H7N9 and A/H3N2 infection to investigate mechanisms underpinning CD38+MHC-II+ phenotype on CD8+ T cells. To further understand MHC-II trogocytosis on murine CD8+ T cells as well as the significance behind the scenario, we used adoptively transferred transgenic OT-I CD8+ T cells and A/H3N2-SIINKEKL infection.
Results
Analysis of influenza-specific immunodominant DbNP366+CD8+ T-cell responses showed that CD38+MHC-II+ co-expression was detected on both virus-specific and bystander CD8+ T cells, with increased numbers of both CD38+MHC-II+CD8+ T-cell populations observed in immune organs including the site of infection during severe viral challenge. OT-I cells adoptively transferred into MHC-II−/− mice had no MHC-II after infection, suggesting that MHC-II was acquired via trogocytosis. The detection of CD19 on CD38+MHC-II+ OT-I cells supports the proposition that MHC-II was acquired by trogocytosis sourced from B cells. Co-expression of CD38+MHC-II+ on CD8+ T cells was needed for optimal recall following secondary infection.
Conclusions
Overall, our study demonstrates that both virus-specific and bystander CD38+MHC-II+ CD8+ T cells are recruited to the site of infection during severe disease, and that MHC-II presence occurs via trogocytosis from antigen-presenting cells. Our findings highlight the importance of the CD38+MHC-II+ phenotype for CD8+ T-cell recall.
Introduction
Influenza virus infections lead to annual seasonal epidemics that result in 243 000–640 000 deaths globally every year.1 In 2017 alone, ˜9.5 million people were hospitalised with lower respiratory tract influenza infections for a total of 81.5 million hospital days.2 Furthermore, disease severity and mortality rapidly escalate when a new influenza A virus (IAV) emerges, for which a matching vaccine is unavailable, as exemplified by the catastrophic 1918–1919 H1N1 pandemic and the emergence of avian-derived H5N1 and H7N9 strains in 20033 and 2013,4 respectively. In the absence of neutralising antibodies, the recall of pre-existing broadly cross-reactive memory CD8+ T cells that recognise conserved peptide epitopes derived from internal viral proteins presented by infected cells can promote recovery against novel influenza A viruses (IAVs) and even different influenza types.5-7 Activated CD8+ T cells can kill virus-infected cells via perforin- and granzyme-mediated lysis but also produce cytokines including IFN-γ and TNF-α, which render the local environment non-permissive for virus replication and ultimately provide some degree of heterosubtypic protection in the antibody-naïve individuals.
The importance of broadly cross-reactive CD8+ T-cell responses in mitigating influenza disease severity has been shown during outbreaks caused by the emergence of novel subtypes, including the 2003 H5N1,8 2009 pandemic H1N17 and 2013 H7N9 strains.9, 10 Our analysis of immune responses in H7N9-infected hospitalised patients in China demonstrated that early proliferation of H7N9-specific CD8+ T cells contributes to disease resolution and survival, as reflected by mild to moderate clinical scores and early time to discharge from hospital.9 In particular, we showed that early, transient co-expression of CD38+ and HLA-DR+ (MHC-II+) on CD8+ T cells during the early stages of H7N9 infection was strongly associated with patient survival.10
Co-expression of CD38, a cyclic ADP ribose hydrolase and cell surface glycoprotein involved in signal transduction, adhesion and calcium signalling,11 and MHC-II on CD8+ T cells is recognised as a classical hallmark of activation during viral infections. CD38+MHC-II+CD8+ T cells can be detected transiently at high levels in patients infected with viruses such as Ebola,12 HCV,13 HIV,14, 15 Dengue,16 pandemic H1N1 IAV17 and more recently SARS-CoV-218, 19 during the acute phase of infection. CD38+MHC-II+CD8+ T cells are known for their proliferative capacity, exhibit effector functions such as cytotoxicity and cytokine production20-22 and typically decrease in frequency following the resolution of infection.18, 21, 22 However, in converse prolonged expression and high frequency of CD38+MHC-II+CD8+ T cells can be associated with a loss of effector function, as shown during chronic viral infections.23, 24 Moreover, H7N9-infected patients with fatal disease outcomes displayed high and prolonged expression of CD38+MHC-II+ on CD8+ T cells, which had minimal capacity to produce IFN-γ, thus linking the prominence of CD38+MHC-II+ phenotype to fatal outcomes in severe IAV-mediated pneumonia.10
Despite the important role of CD38+MHC-II+CD8+ T cells in viral control, the underlying mechanisms driving their activation and regulation during viral infections are still poorly understood. Here, using our established H7N9 and H3N2 influenza wild-type and transgenic mouse models, we determined how disease severity affected the activation of IAV-specific CD38+MHC-II+CD8+ T-cell responses in vivo and the antigenic determinants that drive their activation and expansion. Our study also revealed how this population acquires MHC-II from antigen-presenting cells by trogocytosis and revealed superior recall capacity of CD38+MHC-II+CD8+ T cells.
Results
H7N9 infection in mice induces CD38+MHC-II+PD-1+ activation profiles
Our previous data revealed that patients hospitalised with severe H7N9 influenza had high and prolonged (more than 4 weeks) expression of CD38, MHC-II and PD-1 on CD8+ T cells.10 To understand the mechanisms underlying such hyper-activated CD38, MHC-II and PD-1 phenotype, we first verified the recruitment kinetics and specificity of CD38+MHC-II+ and CD38+PD-1+ CD8+ T cells in a mouse model of H7N9 infection in C57BL/6 (B6) mice. To minimise H7N9-induced mortality, B6 mice were first primed intranasally (i.n.) with 104 EID50 of the avirulent strain A/Chicken/Shanghai/F/98 (H9N2) 60 days prior to i.n. challenge with 106 PFU of A/Shanghai/4664T/2013 (H7N9) virus (Figure 1a). As expected, the majority of CD38+MHC-II+ or DbNP366+CD8+ T cells in MLN and spleen were CD44hi (antigen-experienced), though expression of CD62L, a molecule that mediates trafficking into lymph nodes via high endothelial venules, differed across these two sites (Figure 1b). Overall, ˜20% of CD8+ T cells in the spleen or mediastinal lymph node (MLN) were CD38+MHC-II+ (20.2% for both spleen and MLN) (Figure 1ci and ii, Supplementary figure 1) or CD38+PD-1+ (range 17.8–20.6%) (Supplementary figure 2) on day 10 after H7N9 exposure. Analysis of CD8+ T-cell responses directed at the immunodominant DbNP366 epitope showed 3- to 5-fold enrichment of DbNP366+CD8+ T cells within the CD38+MHC-II+ (Figure 1ci and ii, Supplementary figure 1) or CD38+PD-1+ (Supplementary figures 1 and 2) sets from the spleen and MLN. Such high level of CD38+MHC-II+ or CD38+PD-1+ on tetramer-specific CD8+ T cells after H7N9 infection in mice supports the observations from H7N9 patients.
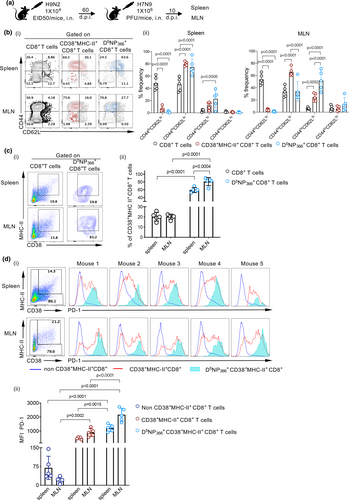
As CD8+ T cells from H7N9-infected hospitalised patients displayed high PD-1 levels, we compared PD-1 expression on H7N9-specific DbNP366+CD38+MHC-II+ CD8+ T cells to that on CD38+MHC-II+ or non-CD38+MHC-II+ CD8+ T cells. DbNP366+CD38+MHC-II+ CD8+ T cells in both the MLN and spleen displayed the highest level of PD-1 (Figure 1d). As expected, PD-1 levels on CD8+ T cells negative for CD38+MHC-II+ were minimal. Thus, high PD-1+ expression on DbNP366+ CD38+MHC-II+ CD8+ and CD38+MHC-II+ CD8+ T cells confirmed the correlation between high CD38+MHC-II+PD-1+ CD8+ T-cell prevalence and severe disease.
Recruitment of CD38+MHC-II+ and CD38+PD-1+CD8+ T cells to the site of infection
To determine how virus load and disease severity affect the recruitment kinetics and phenotype of CD38+MHC-II+ and CD38+PD-1+ CD8+ T cells, we infected B6 mice intranasally with a low (102 PFU) or high (105 PFU) dose of A/HK/x31 (H3N2; X31) virus (Figure 2a). While comparable virus replication kinetics were observed at day 3, 5, 7 and 10 after infection (Figure 2b), the high dose caused more severe disease, as evidenced by significantly greater body weight loss between day 3 and day 7 (P < 0.001) (Figure 2c), and increased lethality (P = 0.003) (Figure 2d) as well as excessive inflammation in the lung (Figure 2e). The 105 PFU X31 virus challenge was associated with more rapid acquisition of a PD-1hi phenotype (P < 0.05 on day 7) on DbNP366+CD8+ T cells (Supplementary figure 3a, b), reflecting the pattern found previously for H7N9-infected mice and patients who succumbed to H7N9 disease.

As expected, both CD38+MHC-II+ and CD38+PD-1+ CD8+ T cells were upregulated during viral challenge and then decreased along with recovery (Figure 2fi, Supplementary figure 3c). These kinetics were also confirmed on adoptively transferred OT-I cells following influenza A virus infection (Supplementary figure 4). Although cell numbers for all CD38+MHC-II+ and CD38+PD-1+ CD8+ T cells in the regional MLN were comparable from day 5 and beyond (Figure 2fi, Supplementary figure 3c), only a small population of cells (11.8%) were DbNP366+ (Figure 2fii, Supplementary figure 3c) within CD38+MHC-II+ CD8+ T cells (peak on day 10) and (17.3%) within CD38+PD-1+CD8+ T cells (peak on day 16), indicating that the majority of CD38+MHC-II+ and CD38+PD-1+ CD8+ T cells recruited during severe influenza disease were tetramer-negative. Furthermore, CD38+MHC-II+ and CD38+PD-1+ CD8+ T cells increased substantially at day 7 at the site of infection (BAL) following the high-dose challenge (Figures 2, 3 and 3, Supplementary figure 3c), while the IAV-specific CD8+ T-cell responses peaked 3 days later (Figures 2fii, and 3, Supplementary figure 3c) and at comparable numbers for both (105 and 102 PFU) groups.
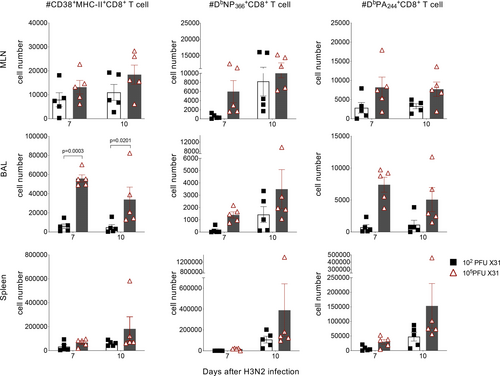
Thus, as a substantially greater number of CD38+MHC-II+ CD8+ T cells was found in comparison to tetramer-specific DbNP366+CD8+ and DbPA224+CD8+ T cells (constituting > 80% of influenza-specific CD8+ T cells25), our findings suggest that a large proportion of CD38+MHC-II+ CD8+ T cell are “bystander” rather than epitope-specific CD8+ T cells. Together, these data suggest that DbNP366 tetramer-negative CD38+MHC-II+CD8+ T cells, many of which numerically will not be virus-specific, extravasate into the lung prior to recruitment of the majority of antigen-specific cells. Whether this is a consequence or a cause of more severe inflammation and pathology is currently unclear.
Murine CD8+ T cells utilise trogocytosis to acquire MHC-II molecule
As expression of the H2-Ab (MHC-II) gene at the transcriptional level depends on the transcription factor CIITA,26, 27 and murine CD8+ T cells lack intrinsically expressed CIITA,28 we further investigated how murine CD8+ T cells acquire expression of MHC-II. We utilised a transgenic OT-I mouse model in the influenza setting. OT-I CD8+ T cells contain a TCRαβ specific for the KbOVA257–264 (OVA; SIINFEKL) epitope, which proliferate after infection with influenza-OVA virus (with SIINFEKL engineered into the neuraminidase stalk).29 We adoptively transferred naïve OT-I CD8+ T cells into MHC-II−/− or C57BL/6 wild-type mice prior to X31-OVA infection (Figure 4a). Greater body weight loss (5%) in MHC-II−/− mice was observed compared to wild-type mice particularly at 3–5 days post-infection (Figure 4b), indicating that lack of MHC-II expression leads to perturbed immune responses and increased disease severity. When we analysed the expansion of OT-I cells (Figure 4c) by frequency (Figure 4d) or number (Figure 4e) of the total CD8+ T cell pool, we observed similar numbers in BAL, MLNs and spleens of both WT and MHC-II−/− mice. As there were also no differences in the combined total number of OT-I cells from these organs of each mouse (Figure 4f), this indicates that OT-I cells proliferate to the same extent in both WT and MHC-II−/− mice.
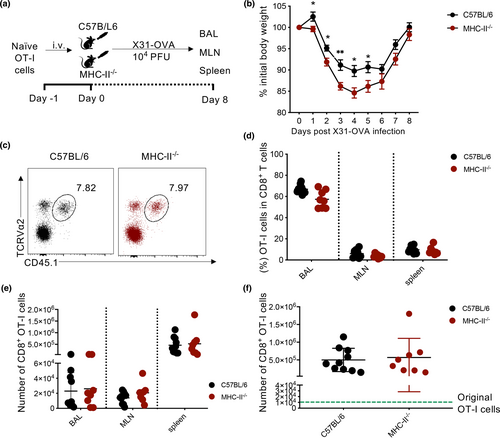
Analysis of MHC-II presence on OT-I cells in BAL, MLNs and spleens of MHC-II−/− or B6 mice showed that MHC-II expression was only limited to OT-I cells transferred into B6 mice and absent in those transferred into MHC-II−/− mice (Figure 5a and b). This observation extended to CD38+ OT-I cells, thus indicating that murine CD8+ T cells do not intrinsically express MHC-II but acquire it from other cells during influenza virus infection.
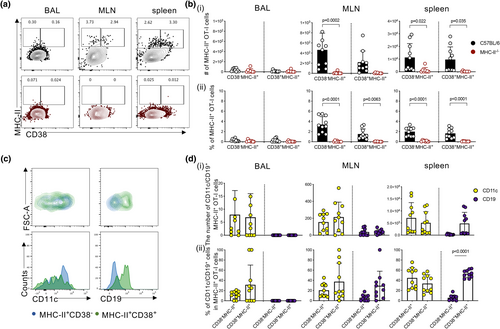
To determine how transferred OT-I cells could have acquired MHC-II, we stained OT-I cells in influenza-infected mice for markers associated with antigen presentation, including CD19 to represent B cells and CD11c as a surrogate DC and macrophage marker.30 In BAL, MLN and spleen, CD11c+ OT-I cells were consistently detected on both CD38−MHC-II+ and CD38+MHC-II+ populations in the BAL (mean 11.7% and 31.4%) as well as the MLN (mean 20.5% vs 37.5%) and spleen (average ˜44.8% vs 33.7%) (Figure 5c and d). In contrast, CD19 was associated mostly with CD38+MHC-II+ OT-I cells (mean 5.7% vs 29.4% in MLN and 5.6% vs 53.0% in spleen), while it was rarely detected on CD38−MHC-II+ and no CD19 expression was observed on OT-I cells in BAL, suggesting that interaction of CD38+MHC-II+CD8+ T cells with CD19+ B cells via trogocytosis provides the source of MHC-II.
To further verify whether the expression of CD19, CD11c and MHC-II was intrinsic to OT-I cells, we measured CD19, CD11c and MHC-II (H2-Ab) mRNA levels in naïve, effector (10 d.p.i.) and memory (60 d.p.i.) CD8+ T-cell populations following X31-OVA infection. Although gene expression of CD11c was not initially detected in naïve populations (Supplementary figure 5a; mean = 14.67), a 2.4 × 102-fold increase was observed in effector (Supplementary figure 5b; mean = 3494) and 1.4 × 102-fold increase in memory (Supplementary figure 5c; mean = 1985.5) CD8+ T-cell populations, demonstrating that CD11c is intrinsic to CD8+ T cells and upregulated as they differentiate. In contrast, this was not the case for CD19 and MHC-II (H2-Ab), with undetectable levels of gene expression observed in naïve, effector and memory CD8+ T-cell populations, similar to levels observed for CD4 gene expression. Overall, our data support the notion that the surface presence of MHC-II on CD8+ T cells is potentially acquired from B cells and dendritic cells/macrophages during influenza virus challenge.
Superior recall capacity of CD38+MHC-II+ CD8+ T cells
To investigate the functional significance of CD38+MHC-II+CD8+ T cells during IAV infection, we analysed the recall capacity of memory OT-I cells established from four subsets of day 8 effectors depicted by CD38 and MHC-II presence (CD38−MHC-II−, CD38+MHC-II−, CD38−MHC-II+, CD38+MHC-II+). OT-I cells adoptively transferred into B6 mice were expanded in vivo following infection with X31-OVA. On day 8 after infection, 4 populations of OT-I cells were FACS-isolated (Figure 6a). Each (CD38−MHC-II−, CD38+MHC-II−, CD38−MHC-II+, CD38+MHC-II+) population was adoptively transferred into separate naive recipient B6 mice and subsequently rested for 30 days to establish memory formation. All mice were then challenged with X31-OVA virus. Mice that received CD38−MHC-II− OT-I cells exhibited significantly (P < 0.05) more body weight loss than mice that received OT-I cells expressing only CD38 (CD38+MHC-II−), MHC-II (CD38−MHC-II+) or both these markers (CD38+MHC-II+) (Figure 6b), indicating an important contribution of these activation markers for protection against severe IAV disease. Importantly, the greatest recall numbers of memory OT-I cells were observed in both the BAL (Figure 6ci and ii) and spleen (Figure 6ci and iii) of mice that received CD38+MHC-II+ cells with OT-I numbers being ˜3.1-fold and ˜4.2-fold higher, respectively, when compared to other subsets.
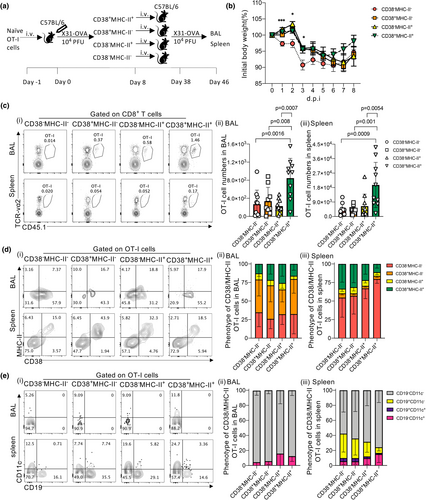
Given that we observed the acquisition of MHC-II molecules by CD8+ T cells during primary infection (Figure 5), we investigated whether this process also occurred following secondary challenge. Irrespective of the original phenotype of OT-I cells transferred, MHC-II+ OT-I cells (mean range of 20.4–31.06% across four groups) were found in BAL (Figure 6di and ii). Furthermore, distinct populations of CD38+MHC-II+ OT-I cells were detected in the spleens of mice across all four groups (Figure 6di and iii), indicating that OT-I cells that did not previously exhibit this phenotype can differentiate into CD38+MHC-II+-expressing cells upon rechallenge. This, however, did not improve their recall ability and highlights the importance of establishing CD38+MHC-II+ phenotype prior to secondary exposure. Interestingly, while both CD19+ and CD11c+ OT-I cells could be found on CD38+MHC-II+ OT-I cells in the spleens of all animal groups (Figure 6ei and iii), only CD19−CD11c+ OT-I cells could be detected in BAL (Figure 6ei and ii). Overall, our findings highlight the importance of the CD38+MHC-II+ phenotype for CD8+ T-cell recall.
Discussion
Co-expression of CD38+MHC-II+ on human CD8+ T cells is a hallmark of viral-specific T cell responses during many viral infections. In particular, early and rapid expansion of CD38+MHC-II+ CD8+ T cells can correlate with effective viraemia control during acute HIV,20 Ebola,12 HCV31 and Dengue16 infection. Our previous studies in H7N9 patients in China showed that the earlier recruitment of activated CD8+ T cells correlated with shorter duration of hospitalisation, while late recruitment of CD8+ T cells leads to more diverse immune responses including upregulation of neutralising antibodies, CD4+ T cells and NK cells.9 While patients who recovered from H7N9 had early IFN-γ-secreting CD8+ T cells,10 persistent and prolonged expression of CD38 and MHC-II on CD8+ T cells was associated with fatal outcomes.10
In this study, we demonstrate that IAV infection in a mouse model also induced the activation and expansion of CD38+MHC-II+ CD8+ T cells. The presence of CD38+ and MHC-II+ on CD8+ T cells recalled following H7N9 challenge correlated with high levels of PD-1 expression in line with our observations in H7N9-infected patients who survived.10 CD38+MHC-II+PD-1+CD8+ T cells were also detected in the primary response following infection with the mouse adapted strain, X31. Of note, increased disease severity resulting from infection with a high dose of X31 was not only characterised by excessive cytokine production, reminiscent of early hypercytokinemia observed during fatal H7N9 infection in humans,9 but also led to increased PD-1+ expression in CD38+MHC-II+CD8+ T cells. This is expected given that CD8+ T-cell exhaustion, defined by PD-1high expression, is a notable feature of disease conditions where excessive or prolonged inflammatory conditions are present, such as during acute and chronic infections caused by highly virulent IAV strains,10 HIV,32, 33 B type hepatitis (HBV)34 and lymphocytic choriomeningitis virus (LCMV).35
Within the total primed CD38+MHC-II+ CD8+ T-cell population in mice following influenza virus infection, a large proportion appeared to be tetramer-negative, which indicates recruitment of bystander CD8+ T cells at the site of infection. This was observed within populations of primary CD38+MHC-II+ CD8+ T-cell populations (Figure 3). This could be because of inflammation-induced expansion not only at the site of infection but also in peripheral immune organs as numerous studies show that cytokines36 such as type I IFN,37 IL-1538 and IL-12/IL-1839 are capable of recruiting non-virus-specific CD8+ T cells during different infectious diseases. It is unclear, however, what impact these bystander CD38+MHC-II+ CD8+ T cells have on the overall immune response against IAV infection. Similar findings have been noted by researchers who found bystander CD8+ T cells activated during the early phases of HIV-1 infection with CD38+MHC-II+ phenotype and specificity for cytomegalovirus, Epstein–Barr virus and influenza antigens.20 Moreover, bystander CD8+ T cells and CD38+MHC-II+ CD8+ T cells can exhibit cytotoxic effects during acute hepatitis A virus infection closely correlating with severe liver injury.40 Thus, while not much is known about the function of bystander CD38+MHC-II+ CD8+ T cells during influenza infection, it is possible that their presence may be related to the hypercytokinemia state at site of infection and contribute to lung damage leading to severe disease outcomes.9 As previous studies suggest that trogocytosis of CD38 can occur during interaction between cells,41 it is also possible that at least some level of CD38 expression could be acquired by trogocytosis.
In contrast to human CD8+ T cells, murine CD8+ T cells lack the transcription factor CIITA, a master regulator of H2-Ab (MHC-II) gene expression.26 However, MHC-II is highly expressed on antigen-presenting cells (APCs) and our results are the first to demonstrate in vivo that CD38+CD8+ T cells can ‘snatch’ MHC-II, along with CD19 and possibly other cell surface markers from B-cell populations. Trogocytosis of cell surface markers has been described for many immune cells in both mice and humans. For example, basophils can acquire MHC-II from dendritic cells to promote T-cell differentiation to Th2 cells.42 Similarly, T cells can acquire HLA-G from APCs changing their function as effector cells to regulatory cells.43 Of relevance, CD38 expressed on B cells can establish lateral associations with various membrane proteins/complexes, including CD19 to help mediate intracellular signalling.44 The interaction of both these markers thus paints a plausible scenario where CD19 is ‘snatched’ from B cells via interaction with CD38 on CD8+ T cells. Recent evidence also indicates that the exchange of markers between CD8+ T-cell and B-cell populations is bi-directional. Strittmatter-Keller et al.45 in particular have reported a population of human B cells expressing CD8 despite the absence of CD8α or CD8β message, suggesting that this marker is acquired via cell-to-cell interactions with T cells. Although CD11c expression was detected on murine MHC-II+CD8+ T cells in our study, it should be noted that murine CD8+ T cells can express a certain level of CD11c intrinsically.
What is to be gained by CD38+CD8+ T cells acquiring MHC-II presence? Our adoptive transfer results demonstrate that MHC-II on murine effector CD38+CD8+ T cells plays a role in their optimal survival and expansion following secondary challenge. In fact, in vitro studies have shown that murine CD8+ T cells that have acquired MHC-II molecules from dendritic cells are able to stimulate CD4+ T cells in culture.46 Given the importance of helper T cells in memory CD8+ T cell formation47, 48 and in the recall of secondary responses,49, 50 it can be assumed that CD8+ T cells expressing MHC-II molecules can also present antigen for recognition by CD4+ T cells to stimulate the secretion of cytokines that promote effector function and differentiation into memory populations. This could therefore constitute an additional mechanism to amplify T-helper responses51 but may also contribute to excessive responses or even tolerance,52 especially if immune-regulatory mechanisms are perturbed as reflected in dysfunctional innate immune responses observed during severe H7N9 infection.9 Other implications can be gleaned from studies demonstrating snatching of peptide-MHC-I complexes (pMHC) by CD8+ T cells from APCs via cell-cell interactions.29, 53, 54 Here, trogocytosis can lead to fratricide of T cells or exhaustion because of prolonged engagement of TCRs with ‘snatched’ MHC-II–antigen complexes especially in the presence of high densities of pMHC.53 These scenarios highlight the impact of trogocytosis of MHC-II molecules, enhancing interaction between CD4+ and CD8+ T cells to boost memory CD8+ T-cell recall responses, but in the event of a severe viral infection, this process may impair CD8+ T-cell responses as observed in patients with fatal H7N9.
Methods
Ethics statement
Animal experiments were conducted according to the Australian National Health and Medical Research Council Code of Practice for the Care and Use of Animals for Scientific Purposes Guidelines for the housing and care of laboratory animals. Ethics was approved by the University of Melbourne Animal Ethics Experimentation Committee (1714108). Animal experiments involving H7N9 infection were approved by the Shanghai Public Health Clinical Center in China (2014-E032-01).
Mice and viral infections
For primary infections, 6- to 8-week-old male or female C57BL/6 mice were lightly anaesthetised with isoflurane and infected by intranasal (i.n.) instillation (30 µL) with 102 and 105 PFU of A/HK/x31 (X31; H3N2). To examine secondary CD8+ T-cell responses in mice following H7N9 infection, mice were first primed i.n. with 104 EID50 of A/Chicken/Shanghai/F/98 (H9N2) virus, followed 60 days later with 106 PFU of A/Shanghai/4664T/2013 (H7N9) virus.
To investigate mechanisms underlying MHC-II presence on murine CD8+ T cells, 104 naïve OT-I cells were adoptively transferred into C57BL/6 mice or MHC-II−/− mice 24 h prior to infection with X31-OVA. After 8 days, CD45.1+CD8+Vα2+ OT-I cells in the BAL, mediastinal lymph node and spleen were assessed. The experiment was repeated twice with 3–5 mice per group for each repeat.
To examine the recall ability of CD38+MHC-II+CD8+ T cells, 104 naïve OT-I cells were adoptively transferred into C57BL/6 mice 24 h prior to infection with X31-OVA. After 8 days, CD45.1+CD8+Vα2+ OT-I cells in the lymph node and spleen were four-way sorted based on their presence of CD38 and MHC-II. Each sorted population was adoptively transferred into separate naïve C57BL/6-recipient mice (5 × 103 cells per mouse) and 30 days later infected with X31-OVA. The experiment was repeated twice with 3–5 mice per group for each repeat.
Tissue sampling and cell preparation
Lungs, spleens, mediastinal lymph nodes (MLN) and bronchoalveolar lavage (BAL) were collected from mice at various time points after infection. Lungs were either homogenised and centrifuged to obtain clarified supernatants to assay for viral titres or enzymatically digested in collagenase III (Worthington Biochemical Corporation, USA; 1 mg mL–1) and DNase I (Sigma-Aldrich, Germany; 0.5 mg mL–1) before passing through cell sieves to obtain single-cell suspensions for analysis. Where necessary, cell suspensions from tissues were incubated with 0.15 m NH4Cl and 17 mm Tris-HCI at pH 7.2 for 5 min at 37°C to lyse red blood cells.
Measurement of lung viral load
Titres of infectious virus were determined by plaque assays on confluent Madin-Darby canine kidney (MDCK) cell monolayers cultured in six-well plates. Cells were infected with lung homogenate supernatants at varying dilutions for 45 min at 37°C before the addition of Leibovitz L15 or MEM medium containing 0.9% agarose overlay containing Trypsin (Worthington Biochemical, NJ, USA), as previously described.55 Plates were incubated at 37°C 5% CO2 for 3 days, and virus-mediated cell lysis then counted as plaques on the cell layer and expressed as plaque forming units (PFU).
Analysis of cytokine levels
Cytokines present in lung homogenate supernatants were measured using a BD CBA flex set (BD Bioscience) as per the manufacturer’s instructions.55 Samples were analysed using a Becton Dickinson FACS Canto II flow cytometer. Data were analysed using FCAP Array software (Soft Flow Inc., Pecs, Hungary).
Tetramer and antibody staining
MHC-I tetramer targeting the immunodominant epitope of the influenza nucleoprotein (DbNP366–374 – ASNENMETM, DbPA224–233 – SSLENFRAYV) was produced in-house and conjugated to streptavidin-APC/PE (Life Technologies Australia Pty Ltd) at a 1:250 dilution at room temperature for 1h. Cells were stained with combinations of fluorochrome-conjugated antibodies: PerCP-Cy5.5-CD3 (#551163), PE-CD8 (#561095), BV421-I-Ab (#562928), APC-CD44 (#553133), FITC-CD44 (#553133), BV711-CD38 (#740697), PerCP-Cy5.5-CD8 (#551162), APC/eF780-CD62L (#47-0621-82), APC-CD11c (#17011481), PE-Cy7-TCR-va2 (#560624) from BD Biosciences, USA; and PE-Cy7-CD38 (#102718), BV785-PD-1 (#329908), APC-Cy7-CD45.1 (#110716), FITC-I-Ab (#116406), Pacific Blue-I-Ab (#116422), FITC-CD19 (#115506) from Biolegend, USA. AF700-CD3 (#56003382) was purchased from Invitrogen, USA.
Live/Dead-aqua 525 was purchased from Invitrogen. Briefly, cell suspensions were stained with Live/Dead Aqua viability dye at room temperature for 10 min followed by staining with tetramer for 15 min and cell surface marker antibodies for 30 min. Cells were fixed with 1% paraformaldehyde before analysis by flow cytometry. All antibody and tetramer staining was performed at 4°C and in the dark. Samples were subsequently acquired on a Becton Dickinson LSR Fortessa or Aria III flow cytometer and data analysed by FlowJo Software (Tree Star Inc., USA).
RNA-Seq
The experiment described in Russ et al.56 was reanalysed for this study. Briefly, RNA from sorted OT-I CTLs (˜106) at naïve, effector and memory stage, respectively, were extracted using TRIzol reagent and purified. RNA-seq libraries were then generated using Illumina’s TruSeq RNA v2 sample preparation protocol according to manufacturers’ instructions, which include cDNA synthesis and cDNA library preparation. PCR amplification on cDNA libraries was performed using Illumina HiSeq2000. RNA-Seq data were aligned to Mouse mm10 genome using HopHat (with Bowtie2) and further analysed using Bioconductor R package, edgeR Bioconductor package.
Statistical analyses
The comparison between absolute numbers of cells was analysed using Tukey's or Sidak's multiple comparisons test. Body weight between mice groups was compared by unpaired t-tests. Absolute numbers of OT-I cells from different mice groups were compared using the Log-rank (Mantel-Cox) test.
Acknowledgments
The authors thank Professor SJ Turner for insightful discussions. This work was supported by the Australian National Health and Medical Research Council (NHMRC) Program Grant (#1071916) and NHMRC Investigator Grant (#1173871) to KK. XJ was a recipient of China Scholarship Council-UoM Joint Scholarship. SYC was a recipient of Melbourne Research Scholarship from University of Melbourne.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
Xiaoxiao Jia: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Writing-original draft. Brendon Y Chua: Formal analysis; Investigation; Writing-original draft. Liyen Loh: Investigation; Supervision; Writing-review & editing. Marios Koutsakos: Investigation; Writing-review & editing. Lukasz Kedzierski: Investigation; Writing-review & editing. Moshe Olshansky: Investigation; Writing-review & editing. William R Heath: Conceptualization; Writing-review & editing. So Young Chang: Data curation; Formal analysis; Writing-review & editing. Jianqing Xu: Conceptualization; Resources; Writing-review & editing. Zhongfang Wang: Conceptualization; Investigation; Supervision; Writing-review & editing. Katherine Kedzierska: Conceptualization; Project administration; Resources; Supervision; Writing-original draft.




