An Efficient Probe for Bacterial Nitroreductase Imaging and Detection Based on NanoLuc-Furimazine Bioluminescent Pair
Ximeng Shi
Jiangsu Key Laboratory for Functional Substances of Chinese Medicine, School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210023 China
These authors contributed equally to this work.
Search for more papers by this authorYumeng Wang
Jiangsu Collaborative Innovation Center of Chinese Medicinal Resources Industrialization, School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210023 China
These authors contributed equally to this work.
Search for more papers by this authorJiaoyang Yu
Clinical Research Center, the Second Hospital of Nanjing, Affiliated Hospital to Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210003 China
Key Laboratory of Resources Biology and Biotechnology in Western China, Ministry of Education, College of Life Sciences, Northwest University, Xi’an, Shaanxi, 710069 China
Search for more papers by this authorYating Yang
Helmholtz International Lab, State Key Laboratory of Microbial Technology, Shandong University, Qingdao, Shandong, 266237 China
Search for more papers by this authorZecheng Jin
Jiangsu Key Laboratory for Functional Substances of Chinese Medicine, School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210023 China
Search for more papers by this authorHanghang Wang
Jiangsu Key Laboratory for Functional Substances of Chinese Medicine, School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210023 China
Search for more papers by this authorDalei Wu
Helmholtz International Lab, State Key Laboratory of Microbial Technology, Shandong University, Qingdao, Shandong, 266237 China
Search for more papers by this authorWei Chen
Clinical Research Center, the Second Hospital of Nanjing, Affiliated Hospital to Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210003 China
Search for more papers by this authorJianming Guo
Jiangsu Collaborative Innovation Center of Chinese Medicinal Resources Industrialization, School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210023 China
Search for more papers by this authorCorresponding Author
Yinan Zhang
Jiangsu Key Laboratory for Functional Substances of Chinese Medicine, School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210023 China
E-mail: [email protected]Search for more papers by this authorXimeng Shi
Jiangsu Key Laboratory for Functional Substances of Chinese Medicine, School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210023 China
These authors contributed equally to this work.
Search for more papers by this authorYumeng Wang
Jiangsu Collaborative Innovation Center of Chinese Medicinal Resources Industrialization, School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210023 China
These authors contributed equally to this work.
Search for more papers by this authorJiaoyang Yu
Clinical Research Center, the Second Hospital of Nanjing, Affiliated Hospital to Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210003 China
Key Laboratory of Resources Biology and Biotechnology in Western China, Ministry of Education, College of Life Sciences, Northwest University, Xi’an, Shaanxi, 710069 China
Search for more papers by this authorYating Yang
Helmholtz International Lab, State Key Laboratory of Microbial Technology, Shandong University, Qingdao, Shandong, 266237 China
Search for more papers by this authorZecheng Jin
Jiangsu Key Laboratory for Functional Substances of Chinese Medicine, School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210023 China
Search for more papers by this authorHanghang Wang
Jiangsu Key Laboratory for Functional Substances of Chinese Medicine, School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210023 China
Search for more papers by this authorDalei Wu
Helmholtz International Lab, State Key Laboratory of Microbial Technology, Shandong University, Qingdao, Shandong, 266237 China
Search for more papers by this authorWei Chen
Clinical Research Center, the Second Hospital of Nanjing, Affiliated Hospital to Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210003 China
Search for more papers by this authorJianming Guo
Jiangsu Collaborative Innovation Center of Chinese Medicinal Resources Industrialization, School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210023 China
Search for more papers by this authorCorresponding Author
Yinan Zhang
Jiangsu Key Laboratory for Functional Substances of Chinese Medicine, School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210023 China
E-mail: [email protected]Search for more papers by this authorComprehensive Summary
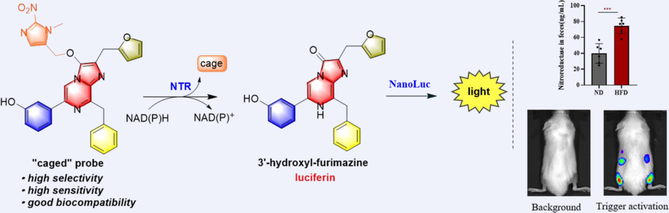
The detection of critical endogenous species, such as bacteria in microenvironments in the body, requires better imaging tools for visualization and monitoring of biological events. Bioluminescence imaging is the most popular strategy for obtaining real-time in living cells and organisms. Herein, we introduced a nitroaryl group on the C-3 position and a hydroxy group at the C-6 phenyl ring on furimazine to report the first bioluminescent probe (7) based on NanoLuc-furimazine bioluminescent pair for the detection of nitroreductase in bacteria. The probe, which possessed up to 560-fold intensity increase with a low detection limit of 16 ng/mL of nitroreductase, has the most efficient uncage efficiency in comparison with other bioluminescent congeners, thus enabling highly selective and sensitive visualization of NTR activity in a panel of clinical priority pathogens. Additionally, imaging of the recombinant strain as well as the NTR from mouse feces indicated the potential of this probe in the application of different mouse disease models.
Supporting Information
| Filename | Description |
|---|---|
| cjoc202300702-sup-0001-Supinfo.pdfPDF document, 4.2 MB |
Appendix S1: Supporting Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1 Kaskova, Z. M.; Tsarkova, A. S.; Yampolsky, I. V. 1001 lights: Luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem. Soc. Rev. 2016, 45, 6048–6077.
- 2 Greer 3rd, L. F.; Szalay, A. A. Imaging of light emission from the expression of luciferases in living cells and organisms: a review. Luminescence 2002, 17, 43–74.
- 3 Prescher, J. A.; Contag, C. H. Guided by the light: visualizing biomolecular processes in living animals with bioluminescence. Curr. Opin. Chem. Biol. 2010, 14, 80–89.
- 4 Welsh, D. K.; Kay, S. A. Bioluminescence imaging in living organisms. Curr. Opin. Biotechnol. 2005, 16, 73–78.
- 5 England, C. G.; Ehlerding, E. B.; Cai, W. NanoLuc: A Small Luciferase Is Brightening Up the Field of Bioluminescence. Bioconjug. Chem. 2016, 27, 1175–1187.
- 6 Iwano, S.; Sugiyama, M.; Hama, H.; Watakabe, A.; Hasegawa, N.; Kuchimaru, T.; Tanaka, K. Z.; Takahashi, M.; Ishida, Y.; Hata, J.; Shimozono, S.; Namiki, K.; Fukano, T.; Kiyama, M.; Okano, H.; Kizaka-Kondoh, S.; McHugh, T. J.; Yamamori, T.; Hioki, H.; Maki, S.; Miyawaki, A. Single-cell bioluminescence imaging of deep tissue in freely moving animals. Science 2018, 359, 935–939.
- 7 Kricka, L. J.; Leach, F.R. In memoriam Dr Marlene DeLuca. 1987 O. M. Smith Lecture. Firefly luciferase: mechanism of action, cloning and expression of the active enzyme. J. Biolumin. Chemilumin. 1989, 3, 1–5.
- 8 Yamaguchi, I. Oplophorus oxyluciferin and a model luciferin compound biologically active with Oplophorus luciferase. Biochem. J. 1975, 151, 9–15.
- 9 Shimomura, O.; Masugi, T.; Johnson, F. H.; Haneda, Y. Properties and reaction mechanism of the bioluminescence system of the deep-sea shrimp Oplophorus gracilorostris. Biochemistry 1978, 17, 994–998.
- 10 Tannous, B. A.; Kim, D. E.; Fernandez, J. L.; Weissleder, R.; Breakefield, X. O. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol. Ther. 2005, 11, 435–443.
- 11 Inouye, S.; Watanabe, K.; Nakamura, H.; Shimomura, O. Secretional luciferase of the luminous shrimp Oplophorus gracilirostris: cDNA cloning of a novel imidazopyrazinone luciferase (1). FEBS Lett. 2000, 481, 19–25.
- 12 Inouye, S.; Sasaki, S. Overexpression, purification and characterization of the catalytic component of Oplophorus luciferase in the deep-sea shrimp, Oplophorus gracilorostris. Protein Express. Purif. 2007, 56, 261–268.
- 13 Hall, M. P.; Unch, J.; Binkowski, B. F.; Valley, M. P.; Butler, B. L.; Wood, M. G.; Otto, P.; Zimmerman, K.; Vidugiris, G.; Machleidt, T.; Robers, M. B.; Benink, H. A.; Eggers, C. T.; Slater, M. R.; Meisenheimer, P. L.; Klaubert, D. H.; Fan, F.; Encell, L. P.; Wood, K. V. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS. Chem. Biol. 2012, 7, 1848–1857.
- 14 Pavlinov, I.; Salkovski, M.; Aldrich, L. N. Beclin 1-ATG14L Protein-Protein Interaction Inhibitor Selectively Inhibits Autophagy through Disruption of VPS34 Complex I. J. Am. Chem. Soc. 2020, 142, 8174–8182.
- 15 Dixon, A. S.; Schwinn, M. K.; Hall, M. P.; Zimmerman, K.; Otto, P.; Lubben, T. H.; Butler, B. L.; Binkowski, B. F.; Machleidt, T.; Kirkland, T. A.; Wood, M. G.; Eggers, C. T.; Encell, L. P.; Wood, K. V. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 2016, 11, 400–408.
- 16 Chen, N.; Cheng, D.; He, T. P.; Yuan, Q. Real-Time Monitoring of Dynamic Chemical Processes in Microbial Metabolism with Optical Sensors. Chin. J. Chem. 2023, 41, 1836–1840.
- 17 Huang, W. J.; Yang, Y. l.; Ye, D. J. Recent Advances in Activatable Probes for Molecular Imaging by Stimuli-Controlled Disassembly. Chin. J. Chem. 2023, 41, 2382–2399.
- 18 Krawczyk, K.; Xue, S.; Buchmann, P.; Charpin-El-Hamri, G.; Saxena, P.; Hussherr, M. D.; Shao, J.; Ye, H.; Xie, M.; Fussenegger, M. Electrogenetic cellular insulin release for real-time glycemic control in type 1 diabetic mice. Science 2020, 368, 993–1001.
- 19 Bryant, D. W.; McCalla, D. R.; Leeksma, M.; Laneuville, P. Type I nitroreductases of Escherichia coli. Can. J. Microbiol. 1981, 27, 81–86.
- 20 Kitamura, S.; Narai, N.; Tatsumi, K. Studies on bacterial nitroreductases. Enzymes involved in reduction of aromatic nitro compounds in Escherichia coli. J. Pharmacobiodyn. 1983, 6, 18–24.
- 21 Roldán, M. D.; Pérez-Reinado, E.; Castillo, F.; Moreno-Vivián, C. Reduction of polynitroaromatic compounds: the bacterial nitroreductases, FEMS. Microbiol. Rev. 2008, 32, 474–500.
- 22 Akiva, E.; Copp, J. N.; Tokuriki, N.; Babbitt, P. C. Evolutionary and molecular foundations of multiple contemporary functions of the nitroreductase superfamily. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E9549–E9558.
- 23 McBain, A. J.; Macfarlane, G. T. Ecological and physiological studies on large intestinal bacteria in relation to production of hydrolytic and reductive enzymes involved in formation of genotoxic metabolites. J. Med. Microbiol. 1998, 47, 407–416.
- 24 Claus, S. P.; Guillou, H.; Ellero-Simatos, S. The gut microbiota: a major player in the toxicity of environmental pollutants. NPJ Biofilms Microbiomes. 2016, 2, 16003.
- 25 Wong, R. H.; Kwong, T.; Yau, K. H.; Au-Yeung, H. Y. Real time detection of live microbes using a highly sensitive bioluminescent nitroreductase probe. Chem. Commun. 2015, 51, 4440–4442.
- 26 Feng, P.; Zhang, H.; Deng, Q.; Liu, W.; Yang, L.; Li, G.; Chen, G.; Du, L.; Ke, B.; Li, M. Real-Time Bioluminescence Imaging of Nitroreductase in Mouse Model. Anal. Chem. 2016, 88, 5610–5614.
- 27 Vorobyeva, A. G.; Stanton, M.; Godinat, A.; Lund, K. B.; Karateev, G. G.; Francis, K. P.; Allen, E.; Gelovani, J. G.; McCormack, E.; Tangney, M; Dubikovskaya, E. A. Development of a Bioluminescent Nitroreductase Probe for Preclinical Imaging. PLoS One 2015, 10, e0131037.
- 28 Yang, X.; Li, Z.; Jiang, T.; Du, L.; Li, M. A coelenterazine-type bioluminescent probe for nitroreductase imaging. Org. Biomol. Chem. 2017, 16, 146–151.
- 29 Mizui, Y.; Eguchi, M.; Tanaka, M.; Ikeda, Y.; Yoshimura, H.; Ozawa, T.; Citterio, D.; Hiruta, Y. Long-term single cell bioluminescence imaging with C-3 position protected coelenterazine analogues. Org. Biomol. Chem. 2021, 19, 579–586.
- 30 Orioka, M.; Eguchi, M.; Mizui, Y.; Ikeda, Y.; Sakama, A.; Li, Q.; Yoshimura, H.; Ozawa, T.; Citterio, D.; Hiruta, Y. A Series of Furimazine Derivatives for Sustained Live-Cell Bioluminescence Imaging and Application to the Monitoring of Myogenesis at the Single-Cell Level. Bioconjug. Chem. 2022, 33, 496–504.
- 31 Li, J.; Chen, L.; Du, L.; Li, M. Cage the firefly luciferin! - a strategy for developing bioluminescent probes. Chem. Soc. Rev. 2013, 42, 662–676.
- 32 Su, T. A.; Bruemmer, K. J.; Chang, C. J. Caged luciferins for bioluminescent activity-based sensing. Curr. Opin. Biotechnol. 2019, 60, 198–204.
- 33 Su, Y.; Walker, J. R.; Park, Y.; Smith, T. P.; Liu, L. X.; Hall, M. P.; Labanieh, L; Hurst, R.; Wang, D. C.; Encell, L. P.; Kim, N.; Zhang, F.; Kay, M. A.; Casey, K. M.; Majzner, R. G.; Cochran, J. R.; Mackall, C. L.; Kirkland, T. A.; Lin, M. Z. Novel NanoLuc substrates enable bright two-population bioluminescence imaging in animals. Nat. Methods 2020, 17, 852–860.
- 34 Haynes, C. A.; Koder, R. L.; Miller, A. F.; Rodgers, D. W. Structures of nitroreductase in three states: effects of inhibitor binding and reduction. J. Biol. Chem. 2002, 277, 11513–11520.
- 35 Wardman, P. Some reactions and properties of nitro radical-anions important in biology and medicine, Environ. Health. Perspect. 1985, 64, 309–320.
- 36 Xu, S.; Wang, Q.; Zhang, Q.; Zhang, L.; Zuo, L.; Jiang, J. D.; Hu, H. Y. Real time detection of ESKAPE pathogens by a nitroreductase-triggered fluorescence turn-on probe. Chem. Commun. 2017, 53, 11177–11180.
- 37 Herrlinger, E. M.; Hau, M.; Redhaber, D. M.; Greve, G.; Willmann, D.; Steimle, S.; Müller, M.; Lübbert, M.; Miething, C. C.; Schüle, R.; Jung, M. Nitroreductase-Mediated Release of Inhibitors of Lysine-Specific Demethylase 1 (LSD1) from Prodrugs in Transfected Acute Myeloid Leukaemia Cells. ChemBioChem 2020, 21, 2329–2347.
- 38 Brennecke, B.; Wang, Q.; Zhang, Q.; Hu, H. Y.; Nazaré, M. An Activatable Lanthanide Luminescent Probe for Time-Gated Detection of Nitroreductase in Live Bacteria. Angew. Chem. Int. Ed. 2020, 59, 8512–8516.
- 39 Wang, Y.; Tong, Q.; Shou, J. W.; Zhao, Z. X.; Li, X.Y.; Zhang, X. F.; Ma, S. R.; He, C. Y.; Lin, Y.; Wen, B. Y.; Guo, F.; Fu, J.; Jiang, J. D. Gut Microbiota-Mediated Personalized Treatment of Hyperlipidemia Using Berberine. Theranostics 2017, 7, 2443–2451.
- 40 Boucher, H. W.; Talbot, G. H.; Bradley, J. S.; Edwards, J. E.; Gilbert, D.; Rice, L. B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12.
- 41 Tomabechi, Y.; Hosoya, T.; Ehara, H.; Sekine, S. I.; Shirouzu, M.; Inouye, S. Crystal structure of nanoKAZ: The mutated 19 kDa component of Oplophorus luciferase catalyzing the bioluminescent reaction with coelenterazine. Biochem. Biophys. Res. Commun. 2016, 470, 88–93.
- 42 Toivonen, J. M.; Boocock, M. R.; Jacobs, H. T. Modelling in Escherichia coli of mutations in mitoribosomal protein S12: novel mutant phenotypes of rpsL. Mol. Microbiol. 1999, 31, 1735–1746.
- 43 Wang, Y.; Wang, Z.; Ji, Q. CRISPR-Cas9-Based Genome Editing and Cytidine Base Editing in Acinetobacter baumannii. STAR Protoc. 2020, 1, 100025.
- 44 Wang, Y.; Wang, S.; Chen, W.; Song, L.; Zhang, Y.; Shen, Z.; Yu, F.; Li, M.; Ji, Q. CRISPR-Cas9 and CRISPR-Assisted Cytidine Deaminase Enable Precise and Efficient Genome Editing in Klebsiella pneumoniae. Appl. Environ. Microbiol. 2018, 84, e01834–18.




