Regulating CRISPR/Cas9 Using Streptavidin-Biotin Interactions†
Wei Shen
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
These authors contribute to this work equally.
†Dedicated to the 130th Anniversary of Wuhan University.
Search for more papers by this authorWei Xiong
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
These authors contribute to this work equally.
†Dedicated to the 130th Anniversary of Wuhan University.
Search for more papers by this authorQianqian Qi
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
These authors contribute to this work equally.
†Dedicated to the 130th Anniversary of Wuhan University.
Search for more papers by this authorXingyu Liu
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorZhongpao Xie
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorYuanyuan Zhang
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorCorresponding Author
Jinxuan Hou
Department of Thyroid & Breast Surgery, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, 430071 China
E-mail: [email protected], [email protected]Search for more papers by this authorCorresponding Author
Tian Tian
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
E-mail: [email protected], [email protected]Search for more papers by this authorXiang Zhou
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorWei Shen
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
These authors contribute to this work equally.
†Dedicated to the 130th Anniversary of Wuhan University.
Search for more papers by this authorWei Xiong
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
These authors contribute to this work equally.
†Dedicated to the 130th Anniversary of Wuhan University.
Search for more papers by this authorQianqian Qi
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
These authors contribute to this work equally.
†Dedicated to the 130th Anniversary of Wuhan University.
Search for more papers by this authorXingyu Liu
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorZhongpao Xie
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorYuanyuan Zhang
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorCorresponding Author
Jinxuan Hou
Department of Thyroid & Breast Surgery, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, 430071 China
E-mail: [email protected], [email protected]Search for more papers by this authorCorresponding Author
Tian Tian
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
E-mail: [email protected], [email protected]Search for more papers by this authorXiang Zhou
College of Chemistry and Molecular Sciences, Wuhan University, Wuhan, Hubei, 430072 China
Search for more papers by this authorComprehensive Summary
Currently, CRISPR/Cas9 technology has found widespread applications across various domains. However, the utility of CRISPR/Cas9 is encumbered by issues pertaining to its reliability and safety, primarily stemming from the uncontrolled activity of the system. Therefore, the design and development of CRISPR/Cas9 systems with controllable activity is of paramount importance. Biotin, characterized by its small molecular weight, and streptavidin, distinguished by its substantial spatial steric hindrance, can be harnessed as an ideal OFF switch (termed a "bioactivity brake") due to their interaction characteristics. In this work, we present a strategy that employs the streptavidin-biotin interaction as a "brake system" for CRISPR/Cas9, effectively allowing for the shutdown of the enzymatic activity of CRISPR/Cas9.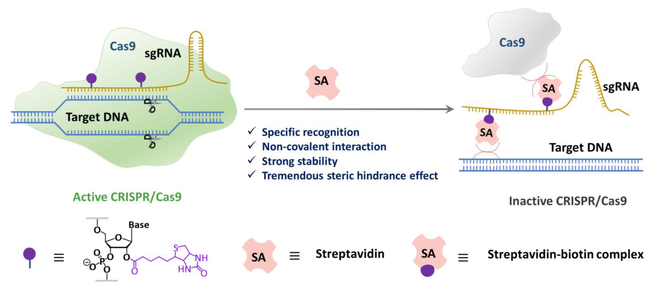
Supporting Information
| Filename | Description |
|---|---|
| cjoc202300662-sup-0001-Supinfo.pdfPDF document, 1.7 MB |
Appendix S1: Supporting Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1 Doudna, J. A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096.
- 2 Cong, L.; Ran, F. A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P. D.; Wu, X.; Jiang, W.; Marraffini, L. A.; Zhang, F. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823.
- 3 Zhu, H.; Li, C.; Gao, C. Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677.
- 4 Hsu, P. D.; Lander, E. S.; Zhang, F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014, 157, 1262–1278.
- 5 Chen, J. S.; Doudna, J. A. The chemistry of Cas9 and its CRISPR colleagues. Nat. Rev. Chem. 2017, 1, 0078.
- 6 Wang, H.; Russa, M. L.; Qi, L. S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264.
- 7 Hsu, P. D.; Scott, D. A.; Weinstein, J. A.; Ran, F. A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E. J.; Wu, X.; Shalem, O.; Cradick, T. J.; Marraffini, L. A.; Bao, G.; Zhang, F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832.
- 8 Lee, H.; Kim, J.-S. Unexpected CRISPR on-target effects. Nat. Biotechnol. 2018, 36, 703–704.
- 9 Pawluk, A.; Amrani, N.; Zhang, Y.; Garcia, B.; Hidalgo-Reyes, Y.; Lee, J.; Edraki, A.; Shah, M.; Sontheimer, E. J.; Maxwell, K. L.; Davidson, A. R. Naturally Occurring Off-Switches for CRISPR-Cas9. Cell 2016, 167, 1829–1838.e1829.
- 10 Harrington, L. B.; Doxzen, K. W.; Ma, E.; Liu, J.-J.; Knott, G. J.; Edraki, A.; Garcia, B.; Amrani, N.; Chen, J. S.; Cofsky, J. C.; Kranzusch, P. J.; Sontheimer, E. J.; Davidson, A. R.; Maxwell, K. L.; Doudna, J. A. A Broad-Spectrum Inhibitor of CRISPR-Cas9. Cell 2017, 170, 1224–1233.e1215.
- 11 Pawluk, A.; Davidson, A. R.; Maxwell, K. L. Anti-CRISPR: discovery, mechanism and function. Nat. Rev. Microbiol. 2018, 16, 12–17.
- 12 Jia, N.; Patel, D. J. Structure-based functional mechanisms and biotechnology applications of anti-CRISPR proteins. Nat. Rev. Mol. Cell Biol. 2021, 22, 563–579.
- 13 Marino, N. D.; Pinilla-Redondo, R.; Csörgő, B.; Bondy-Denomy, J. Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat. Methods 2020, 17, 471–479.
- 14 Maji, B.; Gangopadhyay, S. A.; Lee, M.; Shi, M.; Wu, P.; Heler, R.; Mok, B.; Lim, D.; Siriwardena, S. U.; Paul, B.; Dančík, V.; Vetere, A.; Mesleh, M. F.; Marraffini, L. A.; Liu, D. R.; Clemons, P. A.; Wagner, B. K.; Choudhary, A. A High-Throughput Platform to Identify Small-Molecule Inhibitors of CRISPR-Cas9. Cell 2019, 177, 1067–1079.e1019.
- 15 Lee, S.-W.; Tran, K. T.; Vazquez-Uribe, R.; Gotfredsen, C. H.; Clausen, M. H.; Mendez, B. L.; Montoya, G.; Bach, A.; Sommer, M. O. A. Identification and Optimization of Novel Small-Molecule Cas9 Inhibitors by Cell-Based High-Throughput Screening. J. Med. Chem. 2022, 65, 3266–3305.
- 16 Zhang, Y.; Zhang, Y.; Han, L.; Che, Q.; Tan, J.; Zou, P.; Chen, Y. J. Photo-Modulation of Gene-Editing Enzymes CRISPR/Cas9 with Bifunctional Small-Molecule Ligands. Chin. J. Chem. 2023, 41, 3639–3644.
- 17 Cao, M.; Li, B.; Zhang, X. Anti-CRISPR with non-protein substances. Trends Biotechnol. 2024, 42, 14–16.
- 18 Xiong, W.; Liu, X. Y.; Qi, Q. Q.; Ji, H. M.; Liu, F. B.; Zhong, C.; Liu, S. M.; Tian, T.; Zhou, X. Supramolecular CRISPR-OFF switches with host- guest chemistry. Nucleic Acids Res. 2022, 50, 1241–1255.
- 19 Liu, X. Y.; Xiong, W.; Qi, Q. Q.; Ji, H. M.; Zhang, Y. T.; Lei, H. J.; Liu, J.; Yin, P.; Tian, T.; Zhou, X. A chemical CRISPR off switch efficiently controls gene editing. Cell Rep. Phys. Sci. 2022, 3.
- 20 Liu, X. Y.; Xiong, W.; Qi, Q. Q.; Zhang, Y. T.; Ji, H. M.; Cui, S. Y.; An, J.; Sun, X. M.; Yin, H.; Tian, T.; Zhou, X. Rational guide RNA engineering for small-molecule control of CRISPR/Cas9 and gene editing. Nucleic Acids Res. 2022, 50, 4769–4783.
- 21 Cheng, W.; Liu, F.; Ren, Z.; Chen, W.; Chen, Y.; Liu, T.; Ma, Y.; Cao, N.; Wang, J. Parallel functional assessment of m(6)A sites in human endodermal differentiation with base editor screens. Nat. Commun. 2022, 13, 478.
- 22 Velema, W. A.; Kool, E. T. The chemistry and applications of RNA 2’-OH acylation. Nat. Rev. Chem. 2020, 4, 22–37.
- 23 Kadina, A.; Kietrys, A. M.; Kool, E. T. RNA Cloaking by Reversible Acylation. Angew. Chem. Int. Ed. 2018, 57, 3059–3063.
- 24 Velema, W. A.; Kietrys, A. M.; Kool, E. T. RNA Control by Photoreversible Acylation. J. Am. Chem. Soc. 2018, 140, 3491–3495.
- 25 Velema, W. A.; Park, H. S.; Kadina, A.; Orbai, L.; Kool, E. T. Trapping Transient RNA Complexes by Chemically Reversible Acylation. Angew. Chem. Int. Ed. 2020, 59, 22017–22022.
- 26 Xiao, L.; Fang, L.; Chatterjee, S.; Kool, E. T. Diverse Reagent Scaffolds Provide Differential Selectivity of 2’-OH Acylation in RNA. J. Am. Chem. Soc. 2023, 145, 143–151.
- 27 Wang, S. R.; Wu, L. Y.; Huang, H. Y.; Xiong, W.; Liu, J.; Wei, L.; Yin, P.; Tian, T.; Zhou, X. Conditional control of RNA-guided nucleic acid cleavage and gene editing. Nat. Commun. 2020, 11, 91.
- 28 Gruber, H. J.; Marek, M.; Schindler, H.; Kaiser, K. Biotin−Fluorophore Conjugates with Poly(ethylene glycol) Spacers Retain Intense Fluorescence after Binding to Avidin and Streptavidin. Bioconjugate Chem. 1997, 8, 552–559.
- 29 Ying, L.-Q.; Branchaud, B. P. Design of a reversible biotin analog and applications in protein labeling, detection, and isolation. Chem. Commun. 2011, 47, 8593–8595.
- 30 Sadhu, K. K.; Eierhoff, T.; Römer, W.; Winssinger, N. Photoreductive Uncaging of Fluorophore in Response to Protein Oligomers by Templated Reaction in Vitro and in Cellulo. J. Am. Chem. Soc. 2012, 134, 20013–20016.
- 31 Heinisch, T.; Ward, T. R. Artificial Metalloenzymes Based on the Biotin–Streptavidin Technology: Challenges and Opportunities. Acc. Chem. Res. 2016, 49, 1711–1721.
- 32 Dubacheva, G. V.; Araya-Callis, C.; Geert Volbeda, A.; Fairhead, M.; Codée, J.; Howarth, M.; Richter, R. P. Controlling Multivalent Binding through Surface Chemistry: Model Study on Streptavidin. J. Am. Chem. Soc. 2017, 139, 4157–4167.
- 33 López-Andarias, J.; Saarbach, J.; Moreau, D.; Cheng, Y.; Derivery, E.; Laurent, Q.; González-Gaitán, M.; Winssinger, N.; Sakai, N.; Matile, S. Cell-Penetrating Streptavidin: A General Tool for Bifunctional Delivery with Spatiotemporal Control, Mediated by Transport Systems Such as Adaptive Benzopolysulfane Networks. J. Am. Chem. Soc. 2020, 142, 4784–4792.
- 34 Dundas, C. M.; Demonte, D.; Park, S. Streptavidin–biotin technology: improvements and innovations in chemical and biological applications. Appl. Microbiol. Biotechnol. 2013, 97, 9343–9353.
- 35 Schreiber, C. L.; Smith, B. D. Molecular conjugation using non-covalent click chemistry. Nat. Rev. Chem. 2019, 3, 393–400.




