Unlocking G-Quadruplexes as Targets and Tools against COVID-19
Geng Qin
Laboratory of Chemical Biology and State Key Laboratory of Rare Earth Resource Utilization, Changchun Institute of Applied Chemistry Chinese Academy of Science, Changchun, Jilin, 130022 China
University of Science and Technology of China, Hefei, Anhui, 230026 China
Search for more papers by this authorChuanqi Zhao
Laboratory of Chemical Biology and State Key Laboratory of Rare Earth Resource Utilization, Changchun Institute of Applied Chemistry Chinese Academy of Science, Changchun, Jilin, 130022 China
University of Science and Technology of China, Hefei, Anhui, 230026 China
Search for more papers by this authorJie Yang
Laboratory of Chemical Biology and State Key Laboratory of Rare Earth Resource Utilization, Changchun Institute of Applied Chemistry Chinese Academy of Science, Changchun, Jilin, 130022 China
University of Science and Technology of China, Hefei, Anhui, 230026 China
Search for more papers by this authorZhao Wang
Laboratory of Chemical Biology and State Key Laboratory of Rare Earth Resource Utilization, Changchun Institute of Applied Chemistry Chinese Academy of Science, Changchun, Jilin, 130022 China
University of Science and Technology of China, Hefei, Anhui, 230026 China
Search for more papers by this authorJinsong Ren
Laboratory of Chemical Biology and State Key Laboratory of Rare Earth Resource Utilization, Changchun Institute of Applied Chemistry Chinese Academy of Science, Changchun, Jilin, 130022 China
University of Science and Technology of China, Hefei, Anhui, 230026 China
Search for more papers by this authorCorresponding Author
Xiaogang Qu
Laboratory of Chemical Biology and State Key Laboratory of Rare Earth Resource Utilization, Changchun Institute of Applied Chemistry Chinese Academy of Science, Changchun, Jilin, 130022 China
University of Science and Technology of China, Hefei, Anhui, 230026 China
E-mail: [email protected]Search for more papers by this authorGeng Qin
Laboratory of Chemical Biology and State Key Laboratory of Rare Earth Resource Utilization, Changchun Institute of Applied Chemistry Chinese Academy of Science, Changchun, Jilin, 130022 China
University of Science and Technology of China, Hefei, Anhui, 230026 China
Search for more papers by this authorChuanqi Zhao
Laboratory of Chemical Biology and State Key Laboratory of Rare Earth Resource Utilization, Changchun Institute of Applied Chemistry Chinese Academy of Science, Changchun, Jilin, 130022 China
University of Science and Technology of China, Hefei, Anhui, 230026 China
Search for more papers by this authorJie Yang
Laboratory of Chemical Biology and State Key Laboratory of Rare Earth Resource Utilization, Changchun Institute of Applied Chemistry Chinese Academy of Science, Changchun, Jilin, 130022 China
University of Science and Technology of China, Hefei, Anhui, 230026 China
Search for more papers by this authorZhao Wang
Laboratory of Chemical Biology and State Key Laboratory of Rare Earth Resource Utilization, Changchun Institute of Applied Chemistry Chinese Academy of Science, Changchun, Jilin, 130022 China
University of Science and Technology of China, Hefei, Anhui, 230026 China
Search for more papers by this authorJinsong Ren
Laboratory of Chemical Biology and State Key Laboratory of Rare Earth Resource Utilization, Changchun Institute of Applied Chemistry Chinese Academy of Science, Changchun, Jilin, 130022 China
University of Science and Technology of China, Hefei, Anhui, 230026 China
Search for more papers by this authorCorresponding Author
Xiaogang Qu
Laboratory of Chemical Biology and State Key Laboratory of Rare Earth Resource Utilization, Changchun Institute of Applied Chemistry Chinese Academy of Science, Changchun, Jilin, 130022 China
University of Science and Technology of China, Hefei, Anhui, 230026 China
E-mail: [email protected]Search for more papers by this authorComprehensive Summary
The applicability of G-quadruplexes (G4s) as antiviral targets, therapeutic agents and diagnostic tools for coronavirus disease 2019 (COVID-19) is currently being evaluated, which has drawn the extensive attention of the scientific community. During the COVID-19 pandemic, research in this field is rapidly accumulating. In this review, we summarize the latest achievements and breakthroughs in the use of G4s as antiviral targets, therapeutic agents and diagnostic tools for COVID-19, particularly using G4 ligands. Finally, strength and weakness regarding G4s in anti-SARS-CoV-2 field are highlighted for prospective future projects.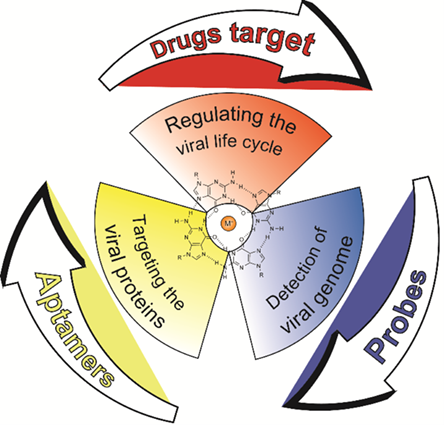
References
- 1 Chan, J. F.; Yuan, S.; Kok, K. H.; To, K. K.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C. C.; Poon, R. W.; Tsoi, H. W.; Lo, S. K.; Chan, K. H.; Poon, V. K.; Chan, W. M.; Ip, J. D.; Cai, J. P.; Cheng, V. C.; Chen, H.; Hui, C. K.; Yuen, K. Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020, 395, 514–523.
- 2 Zhou, P.; Yang, X. L.; Wang, X. G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H. R.; Zhu, Y.; Li, B.; Huang, C. L.; Chen, H. D.; Chen, J.; Luo, Y.; Guo, H.; Jiang, R. D.; Liu, M. Q.; Chen, Y.; Shen, X. R.; Wang, X.; Zheng, X. S.; Zhao, K.; Chen, Q. J.; Deng, F.; Liu, L. L.; Yan, B.; Zhan, F. X.; Wang, Y. Y.; Xiao, G. F.; Shi, Z. L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273.
- 3 Sun, C.; Xie, C.; Bu, G. L.; Zhong, L. Y.; Zeng, M. S. Molecular characteristics, immune evasion, and impact of SARS-CoV-2 variants. Signal. Transduct. Target. Ther. 2022, 7, 202.
- 4 Li, J.; Lai, S.; Gao, G. F.; Shi, W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature 2021, 600, 408–418.
- 5 Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474.
- 6 Guiset Miserachs, H.; Donghi, D.; Borner, R.; Johannsen, S.; Sigel, R. K. Distinct differences in metal ion specificity of RNA and DNA G-quadruplexes. J. Biol. Inorg. Chem. 2016, 21, 975–986.
- 7 Ruggiero, E.; Richter, S. N. G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46, 3270–3283.
- 8 Burge, S.; Parkinson, G. N.; Hazel, P.; Todd, A. K.; Neidle, S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415.
- 9 Fay, M. M.; Lyons, S. M.; Ivanov, P. RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms. J. Mol. Biol. 2017, 429, 2127–2147.
- 10 Brazda, V.; Kolomaznik, J.; Lysek, J.; Bartas, M.; Fojta, M.; Stastny, J.; Mergny, J. L. G4Hunter web application: a web server for G-quadruplex prediction. Bioinformatics 2019, 35, 3493–3495.
- 11 Hon, J.; Martinek, T.; Zendulka, J.; Lexa, M. pqsfinder: an exhaustive and imperfection-tolerant search tool for potential quadruplex- forming sequences in R. Bioinformatics 2017, 33, 3373–3379.
- 12 Kikin, O.; D'Antonio, L.; Bagga, P. S. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006, 34, W676–W682.
- 13 Chambers, V. S.; Marsico, G.; Boutell, J. M.; Di Antonio, M.; Smith, G. P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat .Biotechnol. 2015, 33, 877–881.
- 14 Hansel-Hertsch, R.; Beraldi, D.; Lensing, S. V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; Tannahill, D.; Balasubramanian, S. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272.
- 15 Hansel-Hertsch, R.; Spiegel, J.; Marsico, G.; Tannahill, D.; Balasubramanian, S. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2018, 13, 551–564.
- 16 Yang, S. Y.; Lejault, P.; Chevrier, S.; Boidot, R.; Robertson, A. G.; Wong, J. M. Y.; Monchaud, D. Transcriptome-wide identification of transient RNA G-quadruplexes in human cells. Nat. Commun. 2018, 9, 4730.
- 17 Yang, S. Y.; Monchaud, D.; Wong, J. M. Y. Global mapping of RNA G-quadruplexes (G4-RNAs) using G4RP-seq. Nat. Protoc. 2022, 17, 870–889.
- 18 Dumas, L.; Herviou, P.; Dassi, E.; Cammas, A.; Millevoi, S. G-Quadruplexes in RNA Biology: Recent Advances and Future Directions. Trends Biochem. Sci. 2021, 46, 270–283.
- 19 Hansel-Hertsch, R.; Di Antonio, M.; Balasubramanian, S. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017, 18, 279–284.
- 20 Liu, G.; Du, W.; Xu, H.; Sun, Q.; Tang, D.; Zou, S.; Zhang, Y.; Ma, M.; Zhang, G.; Du, X.; Ju, S.; Cheng, W.; Tian, Y.; Fu, X. RNA G-quadruplex regulates microRNA-26a biogenesis and function. J. Hepatol. 2020, 73, 371–382.
- 21 Maltby, C. J.; Schofield, J. P. R.; Houghton, S. D.; O'Kelly, I.; Vargas-Caballero, M.; Deinhardt, K.; Coldwell, M. J. A 5' UTR GGN repeat controls localisation and translation of a potassium leak channel mRNA through G-quadruplex formation. Nucleic Acids Res. 2020, 48, 9822–9839.
- 22 Mao, S. Q.; Ghanbarian, A. T.; Spiegel, J.; Martinez Cuesta, S.; Beraldi, D.; Di Antonio, M.; Marsico, G.; Hansel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. DNA G-quadruplex structures mold the DNA methylome. Nat. Struct. Mol. Biol. 2018, 25, 951–957.
- 23 Simko, E. A. J.; Liu, H.; Zhang, T.; Velasquez, A.; Teli, S.; Haeusler, A. R.; Wang, J. G-quadruplexes offer a conserved structural motif for NONO recruitment to NEAT1 architectural lncRNA. Nucleic Acids Res. 2020, 48, 7421–7438.
- 24 Weldon, C.; Dacanay, J. G.; Gokhale, V.; Boddupally, P. V. L.; Behm-Ansmant, I.; Burley, G. A.; Branlant, C.; Hurley, L. H.; Dominguez, C.; Eperon, I. C. Specific G-quadruplex ligands modulate the alternative splicing of Bcl-X. Nucleic Acids Res. 2018, 46, 886–896.
- 25 Kosiol, N.; Juranek, S.; Brossart, P.; Heine, A.; Paeschke, K. G-quadruplexes: a promising target for cancer therapy. Mol. Cancer 2021, 20, 40.
- 26 Lejault, P.; Mitteaux, J.; Sperti, F. R.; Monchaud, D. How to untie G-quadruplex knots and why? Cell Chem. Biol. 2021, 28, 436–455.
- 27 Balasubramanian, S.; Hurley, L. H.; Neidle, S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275.
- 28Xu, H.; Di Antonio, M.; McKinney, S.; Mathew, V.; Ho, B.; O'Neil, N. J.; Santos, N. D.; Silvester, J.; Wei, V.; Garcia, J.; Kabeer, F.; Lai, D.; Soriano, P.; Banath, J.; Chiu, D. S.; Yap, D.; Le, D. D.; Ye, F. B.; Zhang, A.; Thu, K.; Soong, J.; Lin, S. C.; Tsai, A. H.; Osako, T.; Algara, T.; Saunders, D. N.; Wong, J.; Xian, J.; Bally, M. B.; Brenton, J. D.; Brown, G. W.; Shah, S. P.; Cescon, D.; Mak, T. W.; Caldas, C.; Stirling, P. C.; Hieter, P.; Balasubramanian, S.; Aparicio, S. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017, 8, 14432.
- 29 Smargiasso, N.; Gabelica, V.; Damblon, C.; Rosu, F.; De Pauw, E.; Teulade-Fichou, M. P.; Rowe, J. A.; Claessens, A. Putative DNA G-quadruplex formation within the promoters of Plasmodium falciparum var genes. BMC Genomics 2009, 10, 362.
- 30 Beaume, N.; Pathak, R.; Yadav, V. K.; Kota, S.; Misra, H. S.; Gautam, H. K.; Chowdhury, S. Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: radioresistance of D. radiodurans involves G4 DNA-mediated regulation. Nucleic Acids Res. 2013, 41, 76–89.
- 31 Perrone, R.; Lavezzo, E.; Riello, E.; Manganelli, R.; Palu, G.; Toppo, S.; Provvedi, R.; Richter, S. N. Mapping and characterization of G-quadruplexes in Mycobacterium tuberculosis gene promoter regions. Sci. Rep. 2017, 7, 5743.
- 32 Abiri, A.; Lavigne, M.; Rezaei, M.; Nikzad, S.; Zare, P.; Mergny, J. L.; Rahimi, H. R. Unlocking G-Quadruplexes as Antiviral Targets. Pharmacol. Rev. 2021, 73, 897–923.
- 33 Hershman, S. G.; Chen, Q.; Lee, J. Y.; Kozak, M. L.; Yue, P.; Wang, L. S.; Johnson, F. B. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008, 36, 144–156.
- 34 Ma, X.; Feng, Y.; Luo, Z.; Zhang, W. Identification and functional characterization of G-quadruplexes in plants. Trends Plant Sci. 2022, 27, 952–953.
- 35 Puig Lombardi, E.; Londono-Vallejo, A.; Nicolas, A. Relationship between G-Quadruplex Sequence Composition in Viruses and Their Hosts. Molecules 2019, 24, 1942.
- 36 Zhai, L. Y.; Liu, J. F.; Zhao, J. J.; Su, A. M.; Xi, X. G.; Hou, X. M. Targeting the RNA G-Quadruplex and Protein Interactome for Antiviral Therapy. J. Med. Chem. 2022, 65, 10161–10182.
- 37 Wang, S. R.; Min, Y. Q.; Wang, J. Q.; Liu, C. X.; Fu, B. S.; Wu, F.; Wu, L. Y.; Qiao, Z. X.; Song, Y. Y.; Xu, G. H.; Wu, Z. G.; Huang, G.; Peng, N. F.; Huang, R.; Mao, W. X.; Peng, S.; Chen, Y. Q.; Zhu, Y.; Tian, T.; Zhang, X. L.; Zhou, X. A highly conserved G-rich consensus sequence in hepatitis C virus core gene represents a new anti-hepatitis C target. Sci. Adv. 2016, 2, e1501535.
- 38 Majee, P.; Pattnaik, A.; Sahoo, B. R.; Shankar, U.; Pattnaik, A. K.; Kumar, A.; Nayak, D. Inhibition of Zika virus replication by G-quadruplex-binding ligands. Mol. Ther. Nucleic Acids 2021, 23, 691–701.
- 39 Zou, M.; Li, J. Y.; Zhang, M. J.; Li, J. H.; Huang, J. T.; You, P. D.; Liu, S. W.; Zhou, C. Q. G-quadruplex binder pyridostatin as an effective multi-target ZIKV inhibitor. Int. J. Biol. Macromol. 2021, 190, 178–188.
- 40 Wang, S. R.; Zhang, Q. Y.; Wang, J. Q.; Ge, X. Y.; Song, Y. Y.; Wang, Y. F.; Li, X. D.; Fu, B. S.; Xu, G. H.; Shu, B.; Gong, P.; Zhang, B.; Tian, T.; Zhou, X. Chemical Targeting of a G-Quadruplex RNA in the Ebola Virus L Gene. Cell Chem. Biol. 2016, 23, 1113–1122.
- 41 Wang, Y.; Grunewald, M.; Perlman, S. Coronaviruses: An Updated Overview of Their Replication and Pathogenesis. Methods Mol. Biol. 2020, 2203, 1–29.
- 42 Huston, N. C.; Wan, H.; Strine, M. S.; de Cesaris Araujo Tavares, R.; Wilen, C. B.; Pyle, A. M. Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Mol. Cell 2021, 81, 584–598.E5.
- 43 Lavezzo, E.; Berselli, M.; Frasson, I.; Perrone, R.; Palu, G.; Brazzale, A. R.; Richter, S. N.; Toppo, S. G-quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide. PLoS Comput. Biol. 2018, 14, e1006675.
- 44 Tan, J.; Vonrhein, C.; Smart, O. S.; Bricogne, G.; Bollati, M.; Kusov, Y.; Hansen, G.; Mesters, J. R.; Schmidt, C. L.; Hilgenfeld, R. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 2009, 5, e1000428.
- 45 Panera, N.; Tozzi, A. E.; Alisi, A. The G-Quadruplex/Helicase World as a Potential Antiviral Approach Against COVID-19. Drugs 2020, 80, 941–946.
- 46 Bartas, M.; Brazda, V.; Bohalova, N.; Cantara, A.; Volna, A.; Stachurova, T.; Malachova, K.; Jagelska, E. B.; Porubiakova, O.; Cerven, J.; Pecinka, P. In-Depth Bioinformatic Analyses of Nidovirales Including Human SARS-CoV-2, SARS-CoV., MERS-CoV Viruses Suggest Important Roles of Non-canonical Nucleic Acid Structures in Their Lifecycles. Front. Microbiol. 2020, 11, 1583.
- 47 Zhang, R.; Xiao, K.; Gu, Y.; Liu, H.; Sun, X. Whole Genome Identification of Potential G-Quadruplexes and Analysis of the G-Quadruplex Binding Domain for SARS-CoV-2. Front. Genet. 2020, 11, 587829.
- 48 Cui, H.; Zhang, L. G-Quadruplexes Are Present in Human Coronaviruses Including SARS-CoV-2. Front. Microbiol. 2020, 11, 567317.
- 49 Ji, D.; Juhas, M.; Tsang, C. M.; Kwok, C. K.; Li, Y.; Zhang, Y. Discovery of G-quadruplex-forming sequences in SARS-CoV-2. Brief Bioinform. 2021, 22, 1150–1160.
- 50 Belmonte-Reche, E.; Serrano-Chacon, I.; Gonzalez, C.; Gallo, J.; Banobre-Lopez, M. Potential G-quadruplexes and i-Motifs in the SARS-CoV-2. PLoS One 2021, 16, e0250654.
- 51 Zhao, C.; Qin, G.; Niu, J.; Wang, Z.; Wang, C.; Ren, J.; Qu, X. Targeting RNA G-Quadruplex in SARS-CoV-2: A Promising Therapeutic Target for COVID-19? Angew. Chem. Int. Ed. 2021, 60, 432–438.
- 52 Qin, G.; Zhao, C.; Liu, Y.; Zhang, C.; Yang, G.; Yang, J.; Wang, Z.; Wang, C. Y.; Tu, C. C.; Guo, Z. D.; Ren, J. S.; Qu, X. G. RNA G-quadruplex formed in SARS-CoV-2 used for COVID-19 treatment in animal models. Cell Discov. 2022, 8, 86.
- 53 Miclot, T.; Hognon, C.; Bignon, E.; Terenzi, A.; Marazzi, M.; Barone, G.; Monari, A. Structure and Dynamics of RNA Guanine Quadruplexes in SARS-CoV-2 Genome. Original Strategies against Emerging Viruses. J. Phys. Chem. Lett. 2021, 12, 10277–10283.
- 54 Oliva, R.; Mukherjee, S.; Manisegaran, M.; Campanile, M.; Del Vecchio, P.; Petraccone, L.; Winter, R. Binding Properties of RNA Quadruplex of SARS-CoV-2 to Berberine Compared to Telomeric DNA Quadruplex. Int. J. Mol. Sci. 2022, 23, 5690.
- 55 Mukherjee, S. K.; Knop, J. M.; Winter, R. Modulation of the Conformational Space of SARS-CoV-2 RNA Quadruplex RG-1 by Cellular Components and the Amyloidogenic Peptides alpha- Synuclein and hIAPP. Chemistry 2022, 28, e202104182.
- 56 Bezzi, G.; Piga, E. J.; Binolfi, A.; Armas, P. CNBP Binds and Unfolds in vitro G-Quadruplexes Formed in the SARS-CoV-2 Positive and Negative Genome Strands. Int. J. Mol. Sci. 2021, 22, 2614.
- 57 Neidle, S. Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010, 277, 1118–1125.
- 58Qiao, J.; Li, Y. S.; Zeng, R.; Liu, F. L.; Luo, R. H.; Huang, C.; Wang, Y. F.; Zhang, J.; Quan, B.; Shen, C.; Mao, X.; Liu, X.; Sun, W.; Yang, W.; Ni, X.; Wang, K.; Xu, L.; Duan, Z. L.; Zou, Q. C.; Zhang, H. L.; Qu, W.; Long, Y. H.; Li, M. H.; Yang, R. C.; Liu, X.; You, J.; Zhou, Y.; Yao, R.; Li, W. P.; Liu, J. M.; Chen, P.; Liu, Y.; Lin, G. F.; Yang, X.; Zou, J.; Li, L.; Hu, Y.; Lu, G. W.; Li, W. M.; Wei, Y. Q.; Zheng, Y. T.; Lei, J.; Yang, S. SARS-CoV-2 M(pro) inhibitors with antiviral activity in a transgenic mouse model. Science 2021, 371, 1374–1378.
- 59 Yang, H.; Rao, Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021, 19, 685–700.
- 60 Zu, S.; Deng, Y. Q.; Zhou, C.; Li, J.; Li, L.; Chen, Q.; Li, X. F.; Zhao, H.; Gold, S.; He, J.; Li, X.; Zhang, C.; Yang, H.; Cheng, G.; Qin, C. F. 25-Hydroxycholesterol is a potent SARS-CoV-2 inhibitor. Cell Res. 2020, 30, 1043–1045.
- 61 Harrison, R. J.; Gowan, S. M.; Kelland, L. R.; Neidle, S. Human telomerase inhibition by substituted acridine derivatives. Bioorg. Med. Chem. Lett. 1999, 9, 2463–2468.
- 62 Burger, A. M.; Dai, F.; Schultes, C. M.; Reszka, A. P.; Moore, M. J.; Double, J. A.; Neidle, S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005, 65, 1489–1496.
- 63 Gowan, S. M.; Harrison, J. R.; Patterson, L.; Valenti, M.; Read, M. A.; Neidle, S.; Kelland, L. R. A G-quadruplex-interactive potent small-molecule inhibitor of telomerase exhibiting in vitro and in vivo antitumor activity. Mol. Pharmacol. 2002, 61, 1154–1162.
- 64 Artusi, S.; Nadai, M.; Perrone, R.; Biasolo, M. A.; Palu, G.; Flamand, L.; Calistri, A.; Richter, S. N. The Herpes Simplex Virus-1 genome contains multiple clusters of repeated G-quadruplex: Implications for the antiviral activity of a G-quadruplex ligand. Antiviral. Res. 2015, 118, 123–131.
- 65 Biswas, B.; Kandpal, M.; Vivekanandan, P. A G-quadruplex motif in an envelope gene promoter regulates transcription and virion secretion in HBV genotype B. Nucleic Acids Res. 2017, 45, 11268–11280.
- 66 Lv, L.; Cui, H.; Chen, Z.; Zhou, Y.; Zhang, L. G-quadruplex ligands inhibit chikungunya virus replication. J. Med. Virol. 2022, 94, 2519–2527.
- 67 Parkinson, G. N.; Ghosh, R.; Neidle, S. Structural basis for binding of porphyrin to human telomeres. Biochemistry 2007, 46, 2390–2397.
- 68 Izbicka, E.; Wheelhouse, R. T.; Raymond, E.; Davidson, K. K.; Lawrence, R. A.; Sun, D.; Windle, B. E.; Hurley, L. H.; Von Hoff, D. D. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 1999, 59, 639–644.
- 69 Grand, C. L.; Han, H.; Munoz, R. M.; Weitman, S.; Von Hoff, D. D.; Hurley, L. H.; Bearss, D. J. The cationic porphyrin TMPyP4 down- regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol. Cancer Ther. 2002, 1, 565–573.
- 70 Shi, D. F.; Wheelhouse, R. T.; Sun, D.; Hurley, L. H. Quadruplex- interactive agents as telomerase inhibitors: synthesis of porphyrins and structure-activity relationship for the inhibition of telomerase. J. Med. Chem. 2001, 44, 4509–4523.
- 71 Piekna-Przybylska, D.; Bambara, R. A.; Maggirwar, S. B.; Dewhurst, S. G-quadruplex ligands targeting telomeres do not inhibit HIV promoter activity and cooperate with latency reversing agents in killing latently infected cells. Cell Cycle 2020, 19, 2298–2313.
- 72Spinner, C. D.; Gottlieb, R. L.; Criner, G. J.; Arribas Lopez, J. R.; Cattelan, A. M.; Soriano Viladomiu, A.; Ogbuagu, O.; Malhotra, P.; Mullane, K. M.; Castagna, A.; Chai, L. Y. A.; Roestenberg, M.; Tsang, O. T. Y.; Bernasconi, E.; Le Turnier, P.; Chang, S. C.; SenGupta, D.; Hyland, R. H.; Osinusi, A. O.; Cao, H.; Blair, C.; Wang, H.; Gaggar, A.; Brainard, D. M.; McPhail, M. J.; Bhagani, S.; Ahn, M. Y.; Sanyal, A. J.; Huhn, G.; Marty, F. M.; GS-US-540-5774 Investigators. Effect of Remdesivir vs Standard Care on Clinical Status at 11 Days in Patients with Moderate COVID-19: A Randomized Clinical Trial. JAMA 2020, 324, 1048–1057.
- 73 Liu, G.; Du, W.; Sang, X.; Tong, Q.; Wang, Y.; Chen, G.; Yuan, Y.; Jiang, L.; Cheng, W.; Liu, D.; Tian, Y.; Fu, X. RNA G-quadruplex in TMPRSS2 reduces SARS-CoV-2 infection. Nat. Commun. 2022, 13, 1444.
- 74 Koirala, D.; Dhakal, S.; Ashbridge, B.; Sannohe, Y.; Rodriguez, R.; Sugiyama, H.; Balasubramanian, S.; Mao, H. A single-molecule platform for investigation of interactions between G-quadruplexes and small-molecule ligands. Nat. Chem. 2011, 3, 782–787.
- 75 Rodriguez, R.; Muller, S.; Yeoman, J. A.; Trentesaux, C.; Riou, J. F.; Balasubramanian, S. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J. Am. Chem. Soc. 2008, 130, 15758–15759.
- 76 Liu, L. Y.; Ma, T. Z.; Zeng, Y. L.; Liu, W.; Mao, Z. W. Structural Basis of Pyridostatin and Its Derivatives Specifically Binding to G-Quadruplexes. J. Am. Chem. Soc. 2022, 144, 11878–11887.
- 77 Kumar, S.; Choudhary, D.; Patra, A.; Bhavesh, N. S.; Vivekanandan, P. Analysis of G-quadruplexes upstream of herpesvirus miRNAs: evidence of G-quadruplex mediated regulation of KSHV miR-K12-1-9,11 cluster and HCMV miR-US33. BMC Mol. Cell Biol. 2020, 21, 67.
- 78 Prado Martins, R.; Findakly, S.; Daskalogianni, C.; Teulade-Fichou, M. P.; Blondel, M.; Fahraeus, R. In Cellulo Protein-mRNA Interaction Assay to Determine the Action of G-Quadruplex-Binding Molecules. Molecules 2018, 23, 3124.
- 79 Lavigne, M.; Helynck, O.; Rigolet, P.; Boudria-Souilah, R.; Nowakowski, M.; Baron, B.; Brule, S.; Hoos, S.; Raynal, B.; Guittat, L.; Beauvineau, C.; Petres, S.; Granzhan, A.; Guillon, J.; Pratviel, G.; Teulade-Fichou, M. P.; England, P.; Mergny, J. L.; Munier-Lehmann, H. SARS-CoV-2 Nsp3 unique domain SUD interacts with guanine quadruplexes and G4-ligands inhibit this interaction. Nucleic Acids Res. 2021, 49, 7695–7712.
- 80Hung, I. F.; Lung, K. C.; Tso, E. Y.; Liu, R.; Chung, T. W.; Chu, M. Y.; Ng, Y. Y.; Lo, J.; Chan, J.; Tam, A. R.; Shum, H. P.; Chan, V.; Wu, A. K.; Sin, K. M.; Leung, W. S.; Law, W. L.; Lung, D. C.; Sin, S.; Yeung, P.; Yip, C. C.; Zhang, R. R.; Fung, A. Y.; Yan, E. Y.; Leung, K. H.; Ip, J. D.; Chu, A. W.; Chan, W. M.; Ng, A. C.; Lee, R.; Fung, K.; Yeung, A.; Wu, T. C.; Chan, J. W.; Yan, W. W.; Chan, W. M.; Chan, J. F.; Lie, A. K.; Tsang, O. T.; Cheng, V. C.; Que, T. L.; Lau, C. S.; Chan, K. H.; To, K. K.; Yuen, K. Y. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet 2020, 395, 1695–1704.
- 81 Unal, M. A.; Bitirim, C. V.; Summak, G. Y.; Bereketoglu, S.; Cevher Zeytin, I.; Besbinar, O.; Gurcan, C.; Aydos, D.; Goksoy, E.; Kocakaya, E.; Eran, Z.; Murat, M.; Demir, N.; Aksoy Ozer, Z. B.; Somers, J.; Demir, E.; Nazir, H.; Ozkan, S. A.; Ozkul, A.; Azap, A.; Yilmazer, A.; Akcali, K. C. Ribavirin shows antiviral activity against SARS-CoV-2 and downregulates the activity of TMPRSS2 and the expression of ACE2 in vitro. Can. J. Physiol. Pharmacol. 2021, 99, 449–460.
- 82 Nishio, M.; Tsukakoshi, K.; Ikebukuro, K. G-quadruplex: Flexible conformational changes by cations, pH, crowding and its applications to biosensing. Biosens. Bioelectron. 2021, 178, 113030.
- 83 Carvalho, J.; Lopes-Nunes, J.; Figueiredo, J.; Santos, T.; Miranda, A.; Riscado, M.; Sousa, F.; Duarte, A. P.; Socorro, S.; Tomaz, C. T.; Felgueiras, M.; Teixeira, R.; Faria, C.; Cruz, C. Molecular Beacon Assay Development for Severe Acute Respiratory Syndrome Coronavirus 2 Detection. Sensors (Basel) 2021, 21, 7015.
- 84 Chen, F.; Li, G.; Wu, C.; Wang, W.; Ma, D. L.; Leung, C. H. A rapid and label-free DNA-based interference reduction nucleic acid amplification strategy for viral RNA detection. Biosens. Bioelectron. 2022, 198, 113829.
- 85 Lee, H.; Lee, H.; Hwang, S. H.; Jeong, W.; Kim, D. E. Detection of SARS-CoV-2 RNA through tandem isothermal gene amplification without reverse transcription. Anal. Chim. Acta 2022, 1212, 339909.
- 86 Zhang, R.; Wu, J.; Ao, H.; Fu, J.; Qiao, B.; Wu, Q.; Ju, H. A Rolling Circle-Amplified G-Quadruplex/Hemin DNAzyme for Chemiluminescence Immunoassay of the SARS-CoV-2 Protein. Anal. Chem. 2021, 93, 9933–9938.
- 87 Gupta, A.; Anand, A.; Jain, N.; Goswami, S.; Anantharaj, A.; Patil, S.; Singh, R.; Kumar, A.; Shrivastava, T.; Bhatnagar, S.; Medigeshi, G. R.; Sharma, T. K.; DBT India Consortium for COVID-19 Research. A novel G-quadruplex aptamer-based spike trimeric antigen test for the detection of SARS-CoV-2. Mol. Ther. Nucleic Acids 2021, 26, 321–332.
- 88 Chou, S. H.; Chin, K. H.; Wang, A. H. DNA aptamers as potential anti-HIV agents. Trends Biochem. Sci. 2005, 30, 231–234.
- 89 Mashima, T.; Matsugami, A.; Nishikawa, F.; Nishikawa, S.; Katahira, M. Unique quadruplex structure and interaction of an RNA aptamer against bovine prion protein. Nucleic Acids Res. 2009, 37, 6249–6258.
- 90 Reyes-Reyes, E. M.; Salipur, F. R.; Shams, M.; Forsthoefel, M. K.; Bates, P. J. Mechanistic studies of anticancer aptamer AS1411 reveal a novel role for nucleolin in regulating Rac1 activation. Mol. Oncol. 2015, 9, 1392–1405.
- 91 Woo, H. M.; Kim, K. S.; Lee, J. M.; Shim, H. S.; Cho, S. J.; Lee, W. K.; Ko, H. W.; Keum, Y. S.; Kim, S. Y.; Pathinayake, P.; Kim, C. J.; Jeong, Y. J. Single-stranded DNA aptamer that specifically binds to the influenza virus NS1 protein suppresses interferon antagonism. Antiviral Res. 2013, 100, 337–345.
- 92 Blaum, B. S.; Wunsche, W.; Benie, A. J.; Kusov, Y.; Peters, H.; Gauss-Muller, V.; Peters, T.; Sczakiel, G. Functional binding of hexanucleotides to 3C protease of hepatitis A virus. Nucleic Acids Res. 2012, 40, 3042–3055.
- 93 Michalowski, D.; Chitima-Matsiga, R.; Held, D. M.; Burke, D. H. Novel bimodular DNA aptamers with guanosine quadruplexes inhibit phylogenetically diverse HIV-1 reverse transcriptases. Nucleic Acids Res. 2008, 36, 7124–7135.
- 94 Ruggiero, E.; Zanin, I.; Terreri, M.; Richter, S. N. G-Quadruplex Targeting in the Fight against Viruses: An Update. Int. J. Mol. Sci. 2021, 22, 10984.
- 95 Artusi, S.; Perrone, R.; Lago, S.; Raffa, P.; Di Iorio, E.; Palu, G.; Richter, S. N. Visualization of DNA G-quadruplexes in herpes simplex virus 1-infected cells. Nucleic Acids Res. 2016, 44, 10343–10353.
- 96 Lista, M. J.; Martins, R. P.; Billant, O.; Contesse, M. A.; Findakly, S.; Pochard, P.; Daskalogianni, C.; Beauvineau, C.; Guetta, C.; Jamin, C.; Teulade-Fichou, M. P.; Fahraeus, R.; Voisset, C.; Blondel, M. Nucleolin directly mediates Epstein-Barr virus immune evasion through binding to G-quadruplexes of EBNA1 mRNA. Nat. Commun. 2017, 8, 16043.
- 97 Murat, P.; Zhong, J.; Lekieffre, L.; Cowieson, N. P.; Clancy, J. L.; Preiss, T.; Balasubramanian, S.; Khanna, R.; Tellam, J. G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat .Chem. Biol. 2014, 10, 358–364.




