Recent Advances in Nanopore Sensing†
Mengjie Cui
Shandong Provincial Key Laboratory of Detection Technology for Tumor Markers, College of Chemistry and Chemical Engineering, Linyi University, Linyi, Shandong, 276005 China
M.C. and Y.G. contributed equally to this work.
Search for more papers by this authorYaxian Ge
Shandong Provincial Key Laboratory of Detection Technology for Tumor Markers, College of Chemistry and Chemical Engineering, Linyi University, Linyi, Shandong, 276005 China
M.C. and Y.G. contributed equally to this work.
Search for more papers by this authorXiao Zhuge
Shandong Provincial Key Laboratory of Detection Technology for Tumor Markers, College of Chemistry and Chemical Engineering, Linyi University, Linyi, Shandong, 276005 China
Search for more papers by this authorXin Zhou
Shandong Provincial Key Laboratory of Detection Technology for Tumor Markers, College of Chemistry and Chemical Engineering, Linyi University, Linyi, Shandong, 276005 China
Search for more papers by this authorCorresponding Author
Dongmei Xi
Shandong Provincial Key Laboratory of Detection Technology for Tumor Markers, College of Chemistry and Chemical Engineering, Linyi University, Linyi, Shandong, 276005 China
*E-mail: [email protected]; [email protected]Search for more papers by this authorCorresponding Author
Shusheng Zhang
Shandong Provincial Key Laboratory of Detection Technology for Tumor Markers, College of Chemistry and Chemical Engineering, Linyi University, Linyi, Shandong, 276005 China
*E-mail: [email protected]; [email protected]Search for more papers by this authorMengjie Cui
Shandong Provincial Key Laboratory of Detection Technology for Tumor Markers, College of Chemistry and Chemical Engineering, Linyi University, Linyi, Shandong, 276005 China
M.C. and Y.G. contributed equally to this work.
Search for more papers by this authorYaxian Ge
Shandong Provincial Key Laboratory of Detection Technology for Tumor Markers, College of Chemistry and Chemical Engineering, Linyi University, Linyi, Shandong, 276005 China
M.C. and Y.G. contributed equally to this work.
Search for more papers by this authorXiao Zhuge
Shandong Provincial Key Laboratory of Detection Technology for Tumor Markers, College of Chemistry and Chemical Engineering, Linyi University, Linyi, Shandong, 276005 China
Search for more papers by this authorXin Zhou
Shandong Provincial Key Laboratory of Detection Technology for Tumor Markers, College of Chemistry and Chemical Engineering, Linyi University, Linyi, Shandong, 276005 China
Search for more papers by this authorCorresponding Author
Dongmei Xi
Shandong Provincial Key Laboratory of Detection Technology for Tumor Markers, College of Chemistry and Chemical Engineering, Linyi University, Linyi, Shandong, 276005 China
*E-mail: [email protected]; [email protected]Search for more papers by this authorCorresponding Author
Shusheng Zhang
Shandong Provincial Key Laboratory of Detection Technology for Tumor Markers, College of Chemistry and Chemical Engineering, Linyi University, Linyi, Shandong, 276005 China
*E-mail: [email protected]; [email protected]Search for more papers by this authorDedicated to the Special Issue of In Situ Target Biomolecule Analysis in Confined Nanospace.
Abstract
Nanopore sensing is developing into a powerful label-free approach to investigate the features of biomolecules at the single-molecule level. When a charged molecule is captured within a nanopore, it modulates the ionic current, which can be recorded in real time to reveal the properties of the target molecule. To date, nanopores have been used to sense a variety of analytes, including DNA, RNA, proteins, enzymes, small molecules, cancer cells, and metal ions, and can also provide information on biomolecular structures. In this review, we highlight the progress made in nanopore sensing over the past five years (2016—2020), and provide an outlook on future developments and directions in the field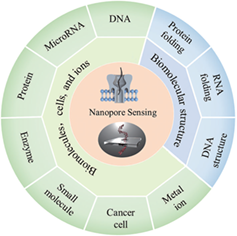
References
- 1 Kasianowicz, J. J.; Brandin, E.; Branton, D.; Deamer, D. W. Characterization of Individual Polynucleotide Molecules Using a Membrane Channel. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 13770–13773.
- 2 Deamer, D.; Akeson, M.; Branton, D. Three Decades of Nanopore Sequencing. Nat. Biotechnol. 2016, 34, 518–524.
- 3 Astier, Y.; Braha, O.; Bayley, H. Toward Single Molecule DNA Sequencing: Direct Identification of Ribonucleoside and Deoxyribonucleoside 5'-Monophosphates by Using an Engineered Protein Nanopore Equipped with a Molecular Adapter. J. Am. Chem. Soc. 2006, 128, 1705–1710.
- 4 Liu, N.; Jiang, Y.; Zhou, Y.; Xia, F.; Guo, W.; Jiang, L. Two-Way Nanopore Sensing of Sequence-Specific Oligonucleotides and Small- Molecule Targets in Complex Matrices Using Integrated DNA Supersandwich Structures. Angew. Chem. Int. Ed. 2013, 52, 2007–2011.
- 5 Wang, Y.; Zheng, D.; Tan, Q.; Wang, M. X.; Gu, L. Q. Nanopore-Based Detection of Circulating MicroRNAs in Lung Cancer Patients. Nat. Nanotechnol. 2011, 6, 668–674.
- 6 Stefureac, R.; Long, Y. T.; Kraatz, H. B.; Howard, P.; Lee, J. S. Transport of Alpha-Helical Peptides through Alpha-Hemolysin and Aerolysin Pores. Biochemistry 2006, 45, 9172–9179.
- 7 Zeng, T.; Li, T.; Li, Y.; Liu, L.; Wang, X.; Liu, Q.; Zhao, Y.; Wu, H. C. DNA-Based Detection of Mercury(II) Ions through Characteristic Current Signals in Nanopores with High Sensitivity and Selectivity. Nanoscale 2014, 6, 8579–8584.
- 8 Wang, L.; Yao, F.; Kang, X. F. Nanopore Single-Molecule Analysis of Metal Ion-Chelator Chemical Reaction. Anal. Chem. 2017, 89, 7958–7965.
- 9 Ding, Y.; Fleming, A. M.; He, L.; Burrows, C. J. Unfolding Kinetics of the Human Telomere I-Motif under a 10 pN Force Imposed by the Alpha-Hemolysin Nanopore Identify Transient Folded-State Lifetimes at Physiological pH. J. Am. Chem. Soc. 2015, 137, 9053–9060.
- 10 Shi, X.; Li, Q.; Gao, R.; Si, W.; Liu, S. C.; Aksimentiev, A.; Long, Y. T. Dynamics of a Molecular Plug Docked onto a Solid-State Nanopore. J. Phys. Chem. Lett. 2018, 9, 4686–4694.
- 11 Liu, H. L.; Jiang, Q. C.; Pang, J.; Jiang, Z. Y.; Cao, J.; Ji, L. N.; Xia, X. H.; Wang, K. A Multiparameter Ph-Sensitive Nanodevice Based on Plasmonic Nanopores. Adv. Funct. Mater. 2018, 28, 7.
- 12 Muthukumar, M.; Plesa, C.; Dekker, C. Single-Molecule Sensing with Nanopores. Phys. Today 2015, 68, 40–46.
- 13 Rauf, S.; Zhang, L.; Ali, A.; Ahmad, J.; Liu, Y.; Li, J. Nanopore-Based, Label-Free, and Real-Time Monitoring Assay for DNA Methyltransferase Activity and Inhibition. Anal. Chem. 2017, 89, 13252–13260.
- 14 Cao, C.; Long, Y. T. Biological Nanopores: Confined Spaces for Electrochemical Single-Molecule Analysis. Acc. Chem. Res. 2018, 51, 331–341.
- 15 Johnson, R. P.; Fleming, A. M.; Beuth, L. R.; Burrows, C. J.; White, H. S. Base Flipping within the Alpha-Hemolysin Latch Allows Single-Molecule Identification of Mismatches in DNA. J. Am. Chem. Soc. 2016, 138, 594–603.
- 16 An, N.; Fleming, A. M.; White, H. S.; Burrows, C. J. Crown Ether-Electrolyte Interactions Permit Nanopore Detection of Individual DNA Abasic Sites in Single Molecules. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 11504–11509.
- 17 Li, W. W.; Gong, L.; Bayley, H. Single-Molecule Detection of 5-Hydroxymethylcytosine in DNA through Chemical Modification and Nanopore Analysis. Angew. Chem. Int. Ed. 2013, 52, 4350–4355.
- 18 Wang, Y.; Zhang, Y.; Guo, Y.; Kang, X. F. Fast and Precise Detection of DNA Methylation with Tetramethylammonium-Filled Nanopore. Sci. Rep. 2017, 7, 183.
- 19 Ayub, M.; Stoddart, D.; Bayley, H. Nucleobase Recognition by Truncated Alpha-Hemolysin Pores. ACS Nano 2015, 9, 7895–7903.
- 20 Ayub, M.; Bayley, H. Engineered Transmembrane Pores. Curr. Opin. Chem. Biol. 2016, 34, 117–126.
- 21 Parker, M. W.; Buckley, J. T.; Postma, J. P.; Tucker, A. D.; Leonard, K.; Pattus, F.; Tsernoglou, D. Structure of the Aeromonas Toxin Proaerolysin in Its Water-Soluble and Membrane-Channel States. Nature 1994, 367, 292–295.
- 22 Cao, C.; Yu, J.; Wang, Y. Q.; Ying, Y. L.; Long, Y. T. Driven Translocation of Polynucleotides through an Aerolysin Nanopore. Anal. Chem. 2016, 88, 5046–5049.
- 23 Cao, C.; Ying, Y. L.; Hu, Z. L.; Liao, D. F.; Tian, H.; Long, Y. T. Discrimination of Oligonucleotides of Different Lengths with a Wild-Type Aerolysin Nanopore. Nat. Nanotechnol. 2016, 11, 713–718.
- 24 Wang, Y. Q.; Li, M. Y.; Qiu, H.; Cao, C.; Wang, M. B.; Wu, X. Y.; Huang, J.; Ying, Y. L.; Long, Y. T. Identification of Essential Sensitive Regions of the Aerolysin Nanopore for Single Oligonucleotide Analysis. Anal. Chem. 2018, 90, 7790–7794.
- 25 Li, M. Y.; Wang, Y. Q.; Lu, Y.; Ying, Y. L.; Long, Y. T. Single Molecule Study of Hydrogen Bond Interactions between Single Oligonucleotide and Aerolysin Sensing Interface. Front. Chem. 2019, 7, 528.
- 26 Wang, L.; Chen, X.; Zhou, S.; Roozbahani, G. M.; Zhang, Y.; Wang, D.; Guan, X. Displacement Chemistry-Based Nanopore Analysis of Nucleic Acids in Complicated Matrices. Chem. Commun. 2018, 54, 13977–13980.
- 27 Xi, D.; Shang, J.; Fan, E.; You, J.; Zhang, S.; Wang, H. Nanopore-Based Selective Discrimination of MicroRNAs with Single-Nucleotide Difference Using Locked Nucleic Acid-Modified Probes. Anal. Chem. 2016, 88, 10540–10546.
- 28 Petersen, M.; Bondensgaard, K.; Wengel, J.; Jacobsen, J. P. Locked Nucleic Acid (LNA) Recognition of RNA: NMR Solution Structures of LNA: RNA Hybrids. J. Am. Chem. Soc. 2002, 124, 5974–5982.
- 29 Wang, S.; Wang, Y.; Yan, S.; Du, X.; Zhang, P.; Chen, H. Y.; Huang, S. Retarded Translocation of Nucleic Acids through Alpha-Hemolysin Nanopore in the Presence of a Calcium Flux. ACS Appl. Mater. Interfaces. 2020, 12, 26926–26935.
- 30 Fang, Z.; Liu, L.; Wang, Y.; Xi, D.; Zhang, S. Unambiguous Discrimination of Multiple Protein Biomarkers by Nanopore Sensing with Double-Stranded DNA-Based Probes. Anal. Chem. 2020, 92, 1730–1737.
- 31 Liu, Q.; Wang, Y.; Liu, Y.; Wang, H.; Li, W.; Tang, P.; Weng, T.; Zhou, S.; Liang, L.; Yuan, J.; Wang, D.; Wang, L. Reduction Chemistry-Assisted Nanopore Determination Method for Immunoglobulin Isotypes. Nanoscale 2020, 12, 19711–19718.
- 32 Guo, B.; Sheng, Y.; Zhou, K.; Liu, Q.; Liu, L.; Wu, H. C. Analyte-Triggered DNA-Probe Release from a Triplex Molecular Beacon for Nanopore Sensing. Angew. Chem. Int. Ed. 2018, 57, 3602–3606.
- 33 Tang, H.; Wang, H.; Yang, C.; Zhao, D.; Qian, Y.; Li, Y. Nanopore-Based Strategy for Selective Detection of Single Carcinoembryonic Antigen (CEA) Molecules. Anal. Chem. 2020, 92, 3042–3049.
- 34 Zhou, S.; Wang, L.; Chen, X.; Guan, X. Label-Free Nanopore Single- Molecule Measurement of Trypsin Activity. ACS Sens. 2016, 1, 607–613.
- 35 Shang, J.; Li, Z.; Liu, L.; Xi, D.; Wang, H. Label-Free Sensing of Human 8-Oxoguanine DNA Glycosylase Activity with a Nanopore. ACS Sens. 2018, 3, 512–518.
- 36 Liu, L.; You, Y.; Zhou, K.; Guo, B.; Cao, Z.; Zhao, Y.; Wu, H. C. A Dual- Response DNA Probe for Simultaneously Monitoring Enzymatic Activity and Environmental pH Using a Nanopore. Angew. Chem. Int. Ed. 2019, 58, 14929–14934.
- 37 Su, Z.; Wei, Y.; Kang, X. F. Simultaneous High-Resolution Detection of Bioenergetic Molecules Using Biomimetic-Receptor Nanopore. Anal. Chem. 2019, 91, 15255–15259.
- 38 Li, X.; Zhai, T.; Gao, P.; Cheng, H.; Hou, R.; Lou, X.; Xia, F. Role of Outer Surface Probes for Regulating Ion Gating of Nanochannels. Nat. Commun. 2018, 9, 40.
- 39 Rauf, S.; Zhang, L.; Ali, A.; Liu, Y.; Li, J. Label-Free Nanopore Biosensor for Rapid and Highly Sensitive Cocaine Detection in Complex Biological Fluids. ACS Sens. 2017, 2, 227–234.
- 40 Yuan, B.; Li, S.; Ying, Y. L.; Long, Y. T. The Analysis of Single Cysteine Molecules with an Aerolysin Nanopore. Analyst 2020, 145, 1179–1183.
- 41 Xi, D.; Li, Z.; Liu, L.; Ai, S.; Zhang, S. Ultrasensitive Detection of Cancer Cells Combining Enzymatic Signal Amplification with an Aerolysin Nanopore. Anal. Chem. 2018, 90, 1029–1034.
- 42 Li, X.; Zhang, P.; Dou, L.; Wang, Y.; Sun, K.; Zhang, X.; Song, G.; Zhao, C.; Li, K.; Bai, Y.; Zeng, X.; Zhou, C.; Ying, B.; Chen, J.; Geng, J. Detection of Circulating Tumor Cells in Breast Cancer Patients by Nanopore Sensing with Aptamer-Mediated Amplification. ACS Sens. 2020, 5, 2359–2366.
- 43 Roozbahani, G. M.; Chen, X.; Zhang, Y.; Wang, L.; Guan, X. Nanopore Detection of Metal Ions: Current Status and Future Directions. Small Methods 2020, 4, 2000266.
- 44 Liu, L.; Fang, Z.; Zheng, X.; Xi, D. Nanopore-Based Strategy for Sensing of Copper(II) Ion and Real-Time Monitoring of a Click Reaction. ACS Sens. 2019, 4, 1323–1328.
- 45 Wei, K.; Yao, F.; Kang, X. F. Single-Molecule Porphyrin-Metal Ion Interaction and Sensing Application. Biosens. Bioelectron. 2018, 109, 272–278.
- 46 Roozbahani, G. M.; Chen, X.; Zhang, Y.; Xie, R.; Ma, R.; Li, D.; Li, H.; Guan, X. Peptide-Mediated Nanopore Detection of Uranyl Ions in Aqueous Media. ACS Sens. 2017, 2, 703–709.
- 47 Wen, S.; Zeng, T.; Liu, L.; Zhao, K.; Zhao, Y.; Liu, X.; Wu, H. C. Highly Sensitive and Selective DNA-Based Detection of Mercury(II) with Alpha-Hemolysin Nanopore. J. Am. Chem. Soc. 2011, 133, 18312–18317.
- 48 Yang, C.; Liu, L.; Zeng, T.; Yang, D.; Yao, Z.; Zhao, Y.; Wu, H. C. Highly Sensitive Simultaneous Detection of Lead(II) and Barium(II) with G-Quadruplex DNA in Alpha-Hemolysin Nanopore. Anal. Chem. 2013, 85, 7302–7307.
- 49 Thoren, K. L.; Worden, E. J.; Yassif, J. M.; Krantz, B. A. Lethal Factor Unfolding Is the Most Force-Dependent Step of Anthrax Toxin Translocation. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 21555–21560.
- 50 Berko, D.; Tabachnick-Cherny, S.; Shental-Bechor, D.; Cascio, P.; Mioletti, S.; Levy, Y.; Admon, A.; Ziv, T.; Tirosh, B.; Goldberg, A. L.; Navon, A. The Direction of Protein Entry into the Proteasome Determines the Variety of Products and Depends on the Force Needed to Unfold Its Two Termini. Mol. Cell 2012, 48, 601–611.
- 51 Oukhaled, G.; Mathe, J.; Biance, A. L.; Bacri, L.; Betton, J. M.; Lairez, D.; Pelta, J.; Auvray, L. Unfolding of Proteins and Long Transient Conformations Detected by Single Nanopore Recording. Phys. Rev. Lett. 2007, 98, 158101.
- 52 Pastoriza-Gallego, M.; Rabah, L.; Gibrat, G.; Thiebot, B.; Van Der Goot, F. G.; Auvray, L.; Betton, J. M.; Pelta, J. Dynamics of Unfolded Protein Transport through an Aerolysin Pore. J. Am. Chem. Soc. 2011, 133, 2923–2931.
- 53 Payet, L.; Martinho, M.; Pastoriza-Gallego, M.; Betton, J. M.; Auvray, L.; Pelta, J.; Mathe, J. Thermal Unfolding of Proteins Probed at the Single Molecule Level Using Nanopores. Anal. Chem. 2012, 84, 4071–4076.
- 54 Si, W.; Aksimentiev, A. Nanopore Sensing of Protein Folding. ACS Nano 2017, 11, 7091–7100.
- 55 Theimer, C. A.; Blois, C. A.; Feigon, J. Structure of the Human Telomerase RNA Pseudoknot Reveals Conserved Tertiary Interactions Essential for Function. Mol. Cell 2005, 17, 671–682.
- 56 Mortimer, S. A.; Kidwell, M. A.; Doudna, J. A. Insights into RNA Structure and Function from Genome-Wide Studies. Nat. Rev. Genet. 2014, 15, 469–479.
- 57 Zhang, X.; Zhang, D.; Zhao, C.; Tian, K.; Shi, R.; Du, X.; Burcke, A. J.; Wang, J.; Chen, S. J.; Gu, L. Q. Nanopore Electric Snapshots of an RNA Tertiary Folding Pathway. Nat. Commun. 2017, 8, 1458.
- 58 Song, L.; Hobaugh, M. R.; Shustak, C.; Cheley, S.; Bayley, H.; Gouaux, J. E. Structure of Staphylococcal Alpha-Hemolysin, a Heptameric Transmembrane Pore. Science 1996, 274, 1859–1866.
- 59 Vercoutere, W.; Winters-Hilt, S.; Olsen, H.; Deamer, D.; Haussler, D.; Akeson, M. Rapid Discrimination among Individual DNA Hairpin Molecules at Single-Nucleotide Resolution Using an Ion Channel. Nat. Biotechnol. 2001, 19, 248–252.
- 60 Vercoutere, W. A.; Winters-Hilt, S.; Deguzman, V. S.; Deamer, D.; Ridino, S. E.; Rodgers, J. T.; Olsen, H. E.; Marziali, A.; Akeson, M. Discrimination among Individual Watson-Crick Base Pairs at the Termini of Single DNA Hairpin Molecules. Nucleic Acids Res. 2003, 31, 1311–1318.
- 61 Sauer-Budge, A. F.; Nyamwanda, J. A.; Lubensky, D. K.; Branton, D. Unzipping Kinetics of Double-Stranded DNA in a Nanopore. Phys. Rev. Lett. 2003, 90, 238101.
- 62 Jin, Q.; Fleming, A. M.; Ding, Y.; Burrows, C. J.; White, H. S. Structural Destabilization of DNA Duplexes Containing Single-Base Lesions Investigated by Nanopore Measurements. Biochemistry 2013, 52, 7870–7877.
- 63 Deguzman, V. S.; Lee, C. C.; Deamer, D. W.; Vercoutere, W. A. Sequence-Dependent Gating of an Ion Channel by DNA Hairpin Molecules. Nucleic Acids Res. 2006, 34, 6425–6437.
- 64 Jin, Q.; Fleming, A. M.; Burrows, C. J.; White, H. S. Unzipping Kinetics of Duplex DNA Containing Oxidized Lesions in an Alpha-Hemolysin Nanopore. J. Am. Chem. Soc. 2012, 134, 11006–11011.
- 65 Schibel, A. E.; Fleming, A. M.; Jin, Q.; An, N.; Liu, J.; Blakemore, C. P.; White, H. S.; Burrows, C. J. Sequence-Specific Single-Molecule Analysis of 8-Oxo-7,8-Dihydroguanine Lesions in DNA Based on Unzipping Kinetics of Complementary Probes in Ion Channel Recordings. J. Am. Chem. Soc. 2011, 133, 14778–14784.
- 66 Perera, R. T.; Fleming, A. M.; Peterson, A. M.; Heemstra, J. M.; Burrows, C. J.; White, H. S. Unzipping of a-Form DNA-RNA, a-Form DNA-PNA, and B-Form DNA-DNA in the Alpha-Hemolysin Nanopore. Biophys. J. 2016, 110, 306–314.
- 67 Liao, D. F.; Cao, C.; Ying, Y. L.; Long, Y. T. A General Strategy of Aerolysin Nanopore Detection for Oligonucleotides with the Secondary Structure. Small 2018, 14, e1704520.
- 68 Lin, Y.; Shi, X.; Liu, S. C.; Ying, Y. L.; Li, Q.; Gao, R.; Fathi, F.; Long, Y. T.; Tian, H. Characterization of DNA Duplex Unzipping through a Sub-2 Nm Solid-State Nanopore. Chem. Commun. 2017, 53, 3539–3542.
- 69 Henderson, E.; Hardin, C. C.; Walk, S. K.; Tinoco, I., Jr.; Blackburn, E. H. Telomeric DNA Oligonucleotides Form Novel Intramolecular Structures Containing Guanine-Guanine Base Pairs. Cell 1987, 51, 899–908.
- 70 Sen, D.; Gilbert, W. Formation of Parallel Four-Stranded Complexes by Guanine-Rich Motifs in DNA and Its Implications for Meiosis. Nature 1988, 334, 364–366.
- 71 Parkinson, G. N.; Lee, M. P.; Neidle, S. Crystal Structure of Parallel Quadruplexes from Human Telomeric DNA. Nature 2002, 417, 876–880.
- 72 Burge, S.; Parkinson, G. N.; Hazel, P.; Todd, A. K.; Neidle, S. Quadruplex DNA: Sequence, Topology and Structure. Nucleic Acids Res. 2006, 34, 5402–5415.
- 73 Wang, Y.; Patel, D. J. Solution Structure of the Human Telomeric Repeat d[AG3(T2AG3)3] G-Tetraplex. Structure 1993, 1, 263–282.
- 74 Lannan, F. M.; Mamajanov, I.; Hud, N. V. Human Telomere Sequence DNA in Water-Free and High-Viscosity Solvents: G-Quadruplex Folding Governed by Kramers Rate Theory. J. Am. Chem. Soc. 2012, 134, 15324–15330.
- 75 Wang, S.; Liang, L.; Tang, J.; Cai, Y.; Zhao, C.; Fang, S.; Wang, H.; Weng, T.; Wang, L.; Wang, D. Label-Free Single-Molecule Identification of Telomere G-Quadruplexes with a Solid-State Nanopore Sensor. RSC Adv. 2020, 10, 27215–27224.
- 76 Zhang, L.; Zhang, K.; Rauf, S.; Dong, D.; Liu, Y.; Li, J. Single-Molecule Analysis of Human Telomere Sequence Interactions with G-Quadruplex Ligand. Anal. Chem. 2016, 88, 4533–4540.




