Unveiling blood pressure-associated genes in aortic cells through integrative analysis of GWAS and RNA modification-associated variants
Huan Zhang, Yuxi Chen, and Peng Xu contributed equally to this study.
Abstract
Background
Genome-wide association studies (GWAS) have identified more than a thousand loci for blood pressure (BP). Functional genes in these loci are cell-type specific. The aim of this study was to elucidate potentially functional genes associated with BP in the aorta through the utilization of RNA modification-associated single-nucleotide polymorphisms (RNAm-SNPs).
Methods
Utilizing large-scale genetic data of 757,601 individuals from the UK Biobank and International Consortium of Blood Pressure consortium, we identified associations between RNAm-SNPs and BP. The association between RNAm-SNPs, gene expression, and BP were examined.
Results
A total of 355 RNAm-SNPs related to m6A, m1A, m5C, m7G, and A-to-I modification were associated with BP. The related genes were enriched in the pancreatic secretion pathway and renin secretion pathway. The BP GWAS signals were significantly enriched with m6A-SNPs, highlighting the potential functional relevance of m6A in physiological processes influencing BP. Notably, m6A-SNPs in CYP11B1, PDE3B, HDAC7, ACE, SLC4A7, PDE1A, FRK, MTHFR, NPPA, CACNA1D, and HDAC9 were identified. Differential methylation and differential expression of the BP genes in FTO-overexpression and METTL14-knockdown vascular smooth muscle cells were detected. RNAm-SNPs were associated with ascending and descending aorta diameter and the genes showed differential methylation between aortic dissection (AD) cases and controls. In scRNA-seq study, we identified ARID5A, HLA-DPB1, HLA-DRA, IRF1, LINC01091, MCL1, MLF1, MLXIPL, NAA16, NADK, RERG, SRM, and USP53 as differential expression genes for AD in aortic cells.
Conclusion
The present study identified RNAm-SNPs in BP loci and elucidated the associations between the RNAm-SNPs, gene expression, and BP. The identified BP-associated genes in aortic cells were associated with AD.
Key points
-
RNA modification-associated single-nucleotide polymorphisms (RNAm-SNPs) related to m6A, m1A, m5C, m7G, and A-to-I modification in blood pressure (BP)-loci were identified.
-
BP Genome-wide association studies (GWAS) signals were significantly enriched with m6A methylation-related SNPs.
-
BP genes exhibited differential methylation and differential expression in FTO-overexpression and METTL14-knockdown vascular smooth muscle cells.
-
BP-genes showed differential methylation and differential expression between aortic dissection cases and controls.
1 INTRODUCTION
Hypertension represents a pervasive global health concern,1 intricately linked to severe conditions such as aortic dissection (AD).2, 3 Although environmental factors play a pivotal role in hypertension onset, genetic determinants are widely acknowledged as crucial contributors to individual blood pressure (BP) regulation.4 Genome-wide association studies (GWAS) have made significant strides in unraveling the genetic basis of hypertension, identifying numerous genomic loci associated with BP.5 However, the majority of these GWAS loci are located in noncoding regions, posing challenges in deciphering their functional implications.
RNA modification, a vital epigenetic process influencing diverse biological regulation processes in living cells,6 encompasses approximately 170 reported types, with N6-methyladenosine (m6A) methylation being extensively studied. Cell-specific patterns characterize RNA modifications,7 offering a unique opportunity to explore the nuanced impact of genetic variants on gene expression within specific cell types. Single-nucleotide polymorphisms (SNPs) associated with RNA modification-associated single-nucleotide polymorphism (RNAm-SNPs) exhibit cell type-specificity,8 making their inclusion in investigations pivotal for uncovering additional layers of cell-specific regulatory mechanisms influencing BP.
Hypertension stands as the foremost risk factor for AD. Aortic cells, closely linked to BP regulation, experience direct mechanical stress induced by high BP. Despite this, functional research on BP-associated genes within aortic tissue is limited. This study delves into the relationship between BP, RNA modification, and gene expression in aortic cells. Emphasis is placed on discerning cell-specific gene expression changes to garner a nuanced understanding of these genes' roles in distinct aorta cell types.
The investigation delineates RNAm-SNPs in BP loci and unveils BP-related functional genes within aortic tissue. The initial step involves identifying RNAm-SNPs within genomic loci linked to BP, followed by assessing their impact on gene expression in aorta cells. Methylation and expression of modifiable genes undergo evaluation in cell experiments. Additionally, Mendelian randomization analysis examines associations between gene expression and BP. To validate our findings, we leverage single-cell transcriptome sequencing data from aortic tissue in cases with AD and matched controls. The cell-specific insights garnered from this study are anticipated to illuminate the intricate cellular mechanisms governing BP regulation.
2 METHODS
2.1 Enrichment of RNAm-SNPs in the BP GWAS data set
In this study, we leveraged innovative RNA modification annotations to elucidate the BP association signals in the 2018 International Consortium of Blood Pressure (ICBP) and UK Biobank GWAS.9 The discovery meta-analysis data sets of this GWAS encompass summary results for associations between approximately 7 million SNPs and systolic blood pressure (SBP) and diastolic blood pressure (DBP). This extensive GWAS includes data from 757,601 individuals, comprising 299,024 participants from the ICBP consortium10, 11 and 458,577 individuals from the UK Biobank.12 The data set can be accessed on the GWAS Catalog website (https://www.ebi.ac.uk/gwas/, accession numbers: GCST006624 and GCST006630).
To identify BP-associated RNAm-SNPs from the vast pool of SNPs in the GWAS data sets, we utilized an annotation file of RNAm-SNPs available in the RMVar database (http://rmvar.renlab.org/download.html). The RMVar database provides comprehensive annotation information for SNPs associated with m6A, m6Am, m1A, m5U, m5C, m7G, 2′-O-Me, A-to-I, and pseudouridine. A total of 1,678,126 SNPs related to these nine types of RNA modifications were annotated, and the data files are downloadable. We integrated the GWAS data files and RNAm-SNP files based on the “rs ID” column using the “merge” function in the R program. This integration annotated GWAS SNPs with RNA modification information, and BP-associated RNAm-SNPs were subsequently selected (considering p < 5.0 × 10−8). Information of the identified RNAm-SNPs was manually cross-verified in NCBI databases. The associations between the identified RNAm-SNPs and BP in East Asian populations were examined in summary data from the Biobank of Japan13 and Taiwan Biobank.14
Within the pool of BP-associated SNPs, we examined whether RNAm-SNPs exhibited overrepresentation compared to what would be expected by chance. A set of non-RNAm-SNPs, equating the number of RNAm-SNPs, was randomly sampled from the GWAS data sets for SBP and DBP. The proportion of SNPs with a p-value < 5.0 × 10−8 in this non-RNAm-SNP set was calculated, repeating this operation 1000 times for each trait to obtain 1000 proportions. The distribution of these proportions served as the background, against which the proportion of RNAm-SNPs with a p-value < 5.0 × 10−8 was compared yielding a p-value.
Furthermore, we applied the fgwas method to assess whether RNAm-SNPs were enriched in GWAS signals for SBP and DBP. This method integrates RNA modification annotation information for each SNP into GWAS summary-level data to evaluate the enrichment of GWAS signals in this annotation type.15 The program was executed following the user manual available at https://github.com/joepickrell/fgwas, with all parameters left at their default settings.
2.2 Cell culture and transfection
To pinpoint RNAm-SNPs associated with BP within distinct m6A methylation loci, we conducted experiments using human aorta smooth muscle cells (HASMCs) subjected to FTO overexpression and METTL14-knockdown. HASMCs, procured from ScienCell (Catalog #6110), were cultured in Smooth Muscle Cell Medium supplemented with 2% fetal bovine serum (Catalog #0010; Thermo Fisher Scientific), 1% smooth muscle cell growth supplement (Catalog #1152), and 1% penicillin/streptomycin solution (Catalog #0503) at 37°C in a humidified 5% CO2 incubator. Media replacement occurred every 48 h, and subculturing was performed at a 1:3 ratio using 0.25% trypsin-EDTA (Gibco, Life Technology) upon reaching subconfluence.
For adenovirus-mediated FTO overexpression, Ad-FTO (Vigenebio) was introduced into the culture medium, with HASMCs transfected at a multiplicity of infection of 100 at 37°C. Ad-GFP, a recombinant adenovirus encoding enhanced GFP, served as a negative control.
2.3 MeRIP-seq and RNA-seq
RNA extraction from FTO overexpression (n = 3), METTL14-knockdown (n = 3), and control (n = 3) HASMCs preceded MeRIP-seq, conducted by Guangzhou Epibiotek Co., Ltd., following our previously published procedure with slight modifications.16 RNA fragmentation into 100 nt fragments, after rRNA removal using a Ribo-Zero rRNA Removal Kit (Illumina; MRZG12324), preceded the construction of a strand-specific RNA library utilizing 10 ng fragmented RNA and the UTP method.
Immunoprecipitation (IP) involved incubating the remaining fragmented RNA with anti-m6A polyclonal antibody (Synaptic Systems; 202003) in IP buffer for 2 h at 4°C. The subsequent IP with protein-A beads (Thermo Fisher Scientific) for an additional 2 h at 4°C was followed by elution of immunoprecipitated RNA from the beads with N6-methyladenosine (Berry & Associates; PR3732) in IP buffer with extraction using TRIzol reagent (Thermo Fisher Scientific; 15596026).
For RNA-seq library generation, the NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina® (NEB; #E7760) utilized purified RNA from both m6A IP and input samples. Sequencing (150 bp paired-end) of the IP library (MeRIP-seq) and input library (RNA-seq) was performed on an Illumina HiSeq. 4000 sequencer (Illumina Inc.).
Quality control, ensuring Q30 > 80%, was executed for paired-end reads obtained from the HiSeq. 4000 sequencer. The resultant qualified reads underwent further processing, including adapter trimming and removal of low-quality reads using Cutadapt software (v1.9.3).17 Subsequent read alignment utilized Hisat2 software (v2.0.4),18 while MACS software19 identified methylated peaks on RNAs. To identify differentially methylated lncRNAs/mRNAs, DiffReps software20 was employed.
2.4 Expression quantitative trait loci (eQTL) analysis for the BP-associated RNAm-SNPs
One of the primary functions of RNA modification is the regulation of gene expression. RNAm-SNPs may influence mRNA expression levels. Therefore, to demonstrate the potential functional relevance of the identified BP-associated RNAm-SNPs, we conducted gene eQTL analysis to explore the associations between RNAm-SNPs and RNA expression levels in the aorta. This analysis was performed using the HaploReg browser (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php).21
2.5 Summary data-based Mendelian randomization (SMR) analysis
We carried out an SMR22 analysis to detect associations between RNA expression levels in the aorta and BP. The SMR software (version 0.712; http://cnsgenomics.com/software/smr/) is a command-line program run on the Windows system. The summary-level data, including SNP rs number, allele information, and summary statistic of the genetic associations, were extracted from the BP GWAS data sets. The binary files containing eQTL summary data from the GTEx project23 were downloaded from http://cnsgenomics.com/software/smr/#DataResource. Additionally, the heterogeneity in dependent instruments (HEIDI) test was performed to discern whether there is a single causal SNP affecting gene expression and BP or if the association is due to linkage disequilibrium. All parameters were maintained at their default setting.
2.6 BP-associated RNAm-SNPs in AD
Aortic aneurysm is characterized by pathological enlargement in the diameter of the aorta. A large-scale GWAS, based on cross-sectional cardiovascular magnetic resonance images in the UK Biobank, identified numerous loci associated with the diameter of the ascending and descending aorta, annotated by a deep learning model.24 This GWAS encompassed 4,374,900 images annotated by a deep learning model from 42,518 participants (40,363 with ascending aorta diameter and 41,415 with descending aorta diameter). We examined the impact of RNAm-SNPs on the diameter of the aortic aorta using data from this study.
Single-cell transcriptome data from a study were utilized to examine the association between gene expression in aortic cell types and AD.25 The study involved six patients and seven healthy controls, and the single-cell gene expression data were retrieved from the GEO database with the accession number GSE207784. The “Seurat” package in R facilitated data integration, filtering, standardization, and quality control. Batch effect correction was performed using the RunHarmony function within the “harmony” R package. Visualization of cell subsets was achieved using Uniform Manifold Approximation and Projection (UMAP). Subsequently, differential expression analysis for each cell type between the cases and controls was conducted using the “muscat” package.26 Genes with low expression levels were filtered out, focusing on those with at least one count in a minimum of 10 cells. In alignment with robust bulk RNA-seq differential expression frameworks, such as edgeR,27 measurements for each sample within each cell type were aggregated using the “muscat” package, resulting in pseudobulk data. This aggregation involved the summation of raw counts. The differential expression analysis was carried out using the edgeR method, as implemented in the “muscat” package. False discovery rates (FDRs) were calculated to adjust for multiple testing (adjusted p-values). Genes exhibiting fold changes (FCs) > 2 and adjusted p-values < 0.05 were considered significant.
Differential m6A between AD cases and controls were examined in data from GSE147028.28
3 RESULTS
3.1 BP-associated RNAm-SNPs
After annotating the GWAS SNPs according to the RNAm-SNPs information, we identified 355 RNAm-SNPs that were significantly associated with BP at p < 5.0 × 10−8. This set comprised 310 m6A-, 22 m1A-, 3 m5C-, 9 m7G-, and 13 A-to-I-related SNPs (Figure 1A). Of these, 350 mapped to 310 known genes (262 protein-coding genes), while 48 mapped to intergenic regions. The 262 protein-coding genes exhibited significant enrichment in pathways related to pancreatic secretion (CELA2A, ADCY9, PLA2G1B, GNAS, ATP2A2, ADCY3, PLCB1, TPCN2, and SLC9A1), the renin secretion pathway (ACE, CREB1, PDE1A, PDE3B, GNAS, CACNA1D, and PLCB1), as well as several immune-related pathways (Figure 1B). Notably, our analysis covered 357 (44.1%) of the 809 BP loci reported in the original ICBP + UKB-GWAS, with RNAm-SNPs identified in 44 loci. It is worth mentioning that the identified RNAm-SNPs did not consistently correspond to the top significant SNPs in these loci.
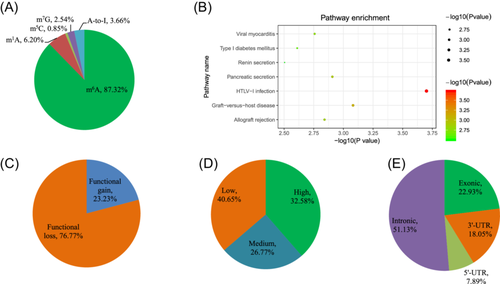
A total of 310 BP-associated m6A-SNPs were identified, with 267 located in protein-coding genes (n = 236) and 49 in long noncoding RNAs and pseudogenes (n = 46). Among them, 72 (23.2%) were functional gain m6A-SNPs and 238 (76.8%) were functional loss m6A-SNPs (Figure 1C). These m6A-SNPs were further categorized as high confidence (32.6%), medium confidence (26.8%), and low confidence (predicted) (40.6%) m6A-SNPs (Figure 1D). Examining the genomic distribution of the 266 protein-coding m6A-SNPs revealed that 136 (51.1%) were intronic, 48 (18.1%) were in the 3′-UTR, 21 (7.9%) were in the 5′-UTR, and 61 (22.9%) were exonic (Figure 1E). A total of 190 unique SBP-associated m6A-SNPs (p < 5.0 × 10−8) were identified (Table 1, Figure 2A, and Supporting Information: Table S1); among these, 146 (66.1%) were classified as high- and medium-confidence. Similarly, we identified 243 DBP-associated m6A-SNPs (Table 1, Figure 2B, and Supporting Information: Table S2), with 171 (62.9%) classified as high- and medium-confidence.
| Total RNAm-SNPs found in GWAS data set | RNAm-SNPs with p < 5.0 × 10−8 (%) | Simulated proportion of genome-wide SNPs with p < 5.0 × 10−8 (95% CI) | p Value | |||||
|---|---|---|---|---|---|---|---|---|
| RNAm | SBP | DBP | SBP | DBP | SBP | DBP | SBP | DBP |
| m6A | 12,276 | 12,391 | 190 (1.55%) | 243 (1.96%) | 0.67%–0.99% | 0.76%–1.11% | 0 | 0 |
| m1A | 653 | 656 | 14 (2.14%) | 16 (2.44%) | 0.61%–1.07% | 0.61%–1.22% | 0 | 0 |
| m7G | 163 | 165 | 7 (4.29%) | 7 (4.24%) | 0.61%–1.23% | 0.61%–1.21% | 0 | 0 |
| A-to-I | 442 | 447 | 6 (1.36%) | 12 (2.68%) | 0.45%–1.13% | 0.67%–1.17% | 2.24E−184 | 0 |
| m5C | 57 | 58 | 2 (5.26%) | 3 (5.17%) | 0.76%–4.59% | 1.72%–3.45% | 1.96E−114 | 8.36E−197 |
| m5U | 3 | 3 | 0 | 1 (33.3%) | - | - | - | - |
| m6Am | 10 | 11 | 0 | 1 (10.0%) | - | - | - | - |
| 2′-O-Me | 4 | 4 | 0 | 0 | - | - | - | - |
| Pseudouridine | 2 | 2 | 0 | 0 | - | - | - | - |
- Abbreviations: CI, confidence interval; DBP, diastolic blood pressure; GWAS, genome-wide association study; RNAm-SNP, RNA modification-associated single-nucleotide polymorphism; SBP, systolic blood pressure.

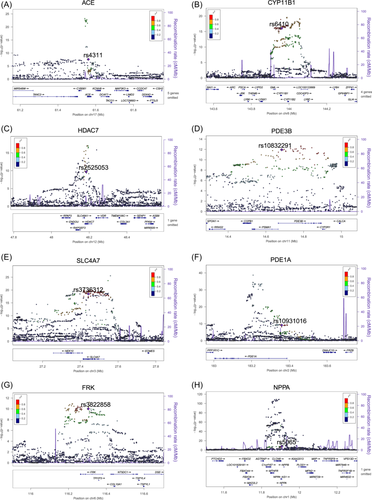
Particularly noteworthy were m6A-SNPs identified in pharmacologically active genes associated with BP. The m6A-SNPs rs4311 in ACE (p = 1.66 × 10−9 and 2.57 × 10−8, respectively) (Figure 3A), rs6410 in CYP11B1 (p = 5.10 × 10−14 and 9.03 × 10−17, respectively) (Figure 3B), rs2525053 in HDAC7 (p = 3.50 × 10−10 and 2.01 × 10−10, respectively) (Figure 3C), rs10832291 in PDE3B (p = 3.60 × 10−9 and 1.16 × 10−12, respectively) (Figure 3D), and rs3736312 in SLC4A7 (p = 3.06 × 10−19 and 3.17 × 10−19, respectively) (Figure 3E) were significantly associated with both SBP and DBP. Additionally, rs10931016 in PDE1A was significantly associated with DBP (p = 7.08 × 10−10) (Figure 3F); rs3822858 in FRK was significantly associated with DBP (p = 9.85 × 10−11) (Figure 3G); rs5065 in the 3′-UTR of NPPA was significantly associated with SBP (p = 1.17 × 10−8) (Figure 3H). Marginal associations included rs5065 in NPPA and DBP (p = 1.17 × 10−7), rs56152532 in CACNA1D and DBP (p = 1.18 × 10−7), and rs2526629 in HDAC9 and SBP (p = 4.45 × 10−6).
We also identified 24 functional loss BP-associated m1A-SNPs, all belonging to the high and medium confidence categories (Supporting Information: Table S3). The m1A-SNP rs7240974 in the 3′-UTR of YES1 was significantly associated with SBP (p = 3.92 × 10−9), with a marginally significant association with DBP (p = 4.87 × 10−7). Nine functional loss m7G-SNPs belonging to the medium confidence category were significantly associated with BP (Supporting Information: Table S4). Thirteen functional loss A-to-I-SNPs belonging to the high confidence category were significantly associated with BP (Supporting Information: Table S5). For m5C modification, three functional loss m5C-SNPs belonging to the high confidence category were significantly associated with BP, including rs113978084 (in the 3′-UTR of LLGL1), rs9986596 (missense variant in ZKSCAN4), and rs10885 (missense variant in PRRC2A). Among the identified BP-associated RNAm-SNPs, 21 of them were associated with BP in East Asian populations (Supporting Information: Table S6).
3.2 Enrichment of RNAm-SNPs in the BP GWAS data set
The proportion of m6A-SNPs, m1A-SNPs, m7G-SNPs, m5C-SNPs, and A-to-I-SNPs with GWAS p-values < 5.0 × 10−8 for SBP and DBP was significantly greater than that of non-RNAm-SNPs (Table 1). Fgwas analysis revealed a notable enrichment of m6A-SNPs among SNPs associated with (p < 5 × 10−8) SBP (log2 enrichment of 2.52, 95% confidence interval [CI]: [1.58, 3.13]), and DBP (log2 enrichment of 2.44, 95% CI: [1.27, 3.10]). This observation underscores the potential functional role of m6A methylation in BP regulation. As a result, we conducted additional experiments to explore the associations between m6A methylation, the expression of these genes, and BP.
3.3 Differentially methylated and expressed RNAs in HASMCs
To elucidate the functional implications of identified BP-associated RNAm-SNPs, we systematically explored their potential impact on gene expression. Our investigative approach encompassed cell culture and transfection experiments, MeRIP-seq, RNA-seq analyses, as well as eQTL and MR analyses. These efforts aimed to unravel the intricate connections between RNAm-SNPs, gene expression, and their ultimate influence on BP regulation.
In FTO-overexpression and METTL14-knockdown HASMCs, we aimed to pinpoint BP loci harboring m6A methylation sites, assess the impact on the expression of methylated genes, and identify BP-associated m6A-SNPs within differentially methylated peaks. Our findings revealed 102 genes with m6A-SNPs that were differentially methylated in FTO-overexpression HASMCs (FC > 2.0, FDR < 0.05) (Supporting Information: Table S7). Additionally, 4 genes (AMOTL2, ZFHX4, UBN1, and JAG1) with m6A-SNPs displayed differential methylation and expression in METTL14-knockdown HASMCs (FC > 2.0, FDR < 0.05) (Supporting Information: Table S8). Notably, 41 of the 102 differentially methylated genes in FTO-overexpression HASMCs exhibited concurrent differential expression. ZFHX4, UBN1, and JAG1 displayed differential methylation in both FTO-overexpression and METTL14-knockdown HASMCs. Among other genes differentially methylated in FTO-overexpression HASMCs, nominal significance in differential methylation was observed for DNAJC11, HDAC7, IPO9, MAP3K1, and STAG3 (FC > 2.0, p < 0.05) (Supporting Information: Table S8). Intriguingly, key BP genes, including MTHFR, HDAC7, and FRK, exhibited differential methylation (Supporting Information: Figure S1), and HDAC7 and FRK showed differential expression in FTO-overexpression HASMCs.
We found that the m6A-SNPs rs4858871, rs3757138, rs1061815, rs330917, rs1147321, rs5870, rs3737058, rs1885987, rs2239925, rs16939357, rs56164415, rs56268858, and rs1051412 situated within differentially methylated peaks in the MAP4, BTN3A2, HLA-A, PPP1R3B, ZBTB6, ACTR1A, AL162274.2, SMG6, NMT1, ZFHX4, BDNF, TBL3, and JAG1 genes, respectively. Of these 13 differentially methylated genes, SMG6 and MAP4 displayed differential expression in FTO-overexpression HASMCs, while ZFHX4 and JAG1 exhibited differential expression in METTL14-knockdown HASMCs.
3.4 Gene expression associated with RNAm-SNPs
Our analysis identified gene expression associations in aorta tissue for 134 BP-associated RNAm-SNPs. These RNAm-SNPs were associated with the expression of 124 genes in the aorta in cis (Supporting Information: Table 1). Notably, rs1879581, rs385691, rs366858, rs76324150, rs17650901, rs7350928 and rs17574425 were associated with expression levels of CRHR1; rs385691, rs366858, rs76324150, rs17650901, rs7350928, and rs17574425 were associated with expression levels of MAPT; rs5065 in the 3′-UTR of NPPA demonstrated an association with expression levels of NPPA-AS1 in the aorta.
3.5 Gene expression associated with BP
In SMR analysis, we detected 38 significant associations involving 29 genes (pSMR < 5.0 × 10−6) (Supporting Information: Table S10). In the eQTL analysis, we identified the association between RNAm-SNPs and gene expression; in SMR analysis, we showed that the gene expression was associated with BP. Notably, for the 41 differentially methylated and differentially expressed genes, the expression levels of BTN3A2, CCHCR1, CDK2AP1, DDI2, GIGYF1, HLA-A, MICB, MRAS, SPATA2L, and UBN1 in the aorta were associated with BP in the SMR analysis.
3.6 RNAm-SNPs and the thoracic aorta
Furthermore, we identified 25 and 17 RNAm-SNPs associated with ascending and descending aorta diameter at p < 5 × 10−6, respectively (Supporting Information: Table S11). Notably, five aorta diameter-associated RNAm-SNPs exhibited cis-eQTL signals in aorta tissues, including rs6673179 in CCBL2, rs2979247 in ERI1, rs1678 in NOC3L, rs12889267 in ARHGEF40, and rs12450028 SRR (Supporting Information: Table S12). SRR demonstrated a significant association with the diameter of the ascending aorta (pSMR = 1.55 × 10−8), and CCBL2 exhibited a nominal association with the ascending aorta diameter (pSMR = 2.01 × 10−5).
Based on the results of the differential m6A study (GSE147028), 42 genes with BP-associated RNAm-SNPs exhibited differential methylation between AD cases and controls (Supporting Information: Table S13). Subsequent scRNA-seq analysis revealed differential expression genes in diverse cell types of the aorta tissue for AD (Figure 4A). In the scRNA-seq study, we identified 328 differential expression genes in 8 cell types (Figure 4B, Supporting Information: Table S14). Among them, ARID5A, HLA-DPB1, HLA-DRA, IRF1, LINC01091, MCL1, MLF1, MLXIPL, NAA16, NADK, RERG, SRM, and USP53 harbored BP-associated RNAm-SNPs. Specifically, HLA-DRA, IRF1, RERG, SRM, and USP53 were identified as differential m6A genes in AD (Table 2).

| SNP | Modification type | GWAS p-value | Trait | Gene | Differential expression in single-cells | Differential m6A methylation | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell type | log2FC | p Value | Adjusted p-value | log2FC | p Value | Adjusted p-value | |||||
| rs1051336 | m7G | 2.51E−18 | DBP | HLA-DRA | Macrophage | −3.18 | 1.96E−05 | 9.18E−03 | 2.17 | 4.68E−06 | 2.14E−04 |
| rs11242115 | m6A | 8.19E−18 | DBP | IRF1 | Fibroblast | −2.40 | 1.59E−04 | 2.93E−02 | 1.41 | 6.47E−06 | 2.76E−04 |
| rs2070725 | m6A | 1.46E−08 | SBP | IRF1 | Fibroblast | −2.40 | 1.59E−04 | 2.93E−02 | 1.41 | 6.47E−06 | 2.76E−04 |
| rs2070725 | m6A | 3.64E−14 | DBP | IRF1 | Fibroblast | −2.40 | 1.59E−04 | 2.93E−02 | 1.41 | 6.47E−06 | 2.76E−04 |
| rs7962491 | m6A | 3.06E−08 | DBP | RERG | Fibroblast | −1.24 | 5.22E−04 | 4.59E−02 | −2.42 | 1.69E−06 | 9.36E−05 |
| rs1884429 | m6A | 4.02E−06 | SBP | SRM | Fibroblast | 1.58 | 6.50E−04 | 4.91E−02 | 1.28 | 4.44E−04 | 7.66E−03 |
| rs79157079 | m6A | 2.37E−08 | SBP | USP53 | Neuronal | 2.19 | 3.03E−03 | 4.02E−02 | −1.57 | 3.48E−07 | 2.56E−05 |
| rs3749591 | m6A | 5.69E−07 | DBP | USP53 | Neuronal | 2.19 | 3.03E−03 | 4.02E−02 | −1.57 | 3.48E−07 | 2.56E−05 |
- Abbreviations: DBP, diastolic blood pressure; FC, fold change; GWAS, genome-wide association study; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
4 DISCUSSION
In this study, our comprehensive analysis of BP-associated RNAm-SNPs provides valuable insights into the genetic and epigenetic landscape of BP regulation. The integration of GWAS data with RNAm-SNPs annotation revealed RNAm-SNPs significantly associated with BP, encompassing various RNA modifications, m6A, m1A, m5C, m7G, and A-to-I modification. It is suggested that RNA modification, especially m6A methylation, may play a role in BP regulation, as the enrichment analysis showed that GWAS signals were significantly enriched with m6A-SNPs. These SNPs were associated with gene expression levels that were related to BP. The results suggested that the RNAm-SNPs affect gene expression controlled by RNA modification in the aorta and the altered mRNA expression levels may result in AD. Our findings contribute to the understanding of the genetic basis of BP regulation, shedding light on the intricate connections between RNA modifications and BP-associated genomic loci.
The overrepresentation of RNAm-SNPs prompted further investigation into potential functional mechanisms. To delve into the regulatory landscape, we explored the differential m6A methylation sites in FTO overexpression and METTL14-knockdown HASMCs. The identification of genes, including HDAC7, FRK, and MTHFR, exhibiting both differential methylation and expression in FTO-overexpression HASMCs, provides a foundation for understanding the functional consequences of m6A modifications in BP-associated genes. To strengthen the functional relevance of identified RNAm-SNPs, we conducted eQTL analysis in aortic tissues. The associations between RNAm-SNPs and RNA expression levels underscore the potential regulatory impact of RNA modifications on gene expression in relevant physiological contexts. Expanding our investigation to AD and single-cell transcriptome data provided additional dimensions to our study. The examination of RNAm-SNPs in the context of AD highlighted potential associations with pathologic enlargement in the diameter of the aorta. Single-cell transcriptomics offered insights into cell-specific gene expression patterns shedding light on the cellular heterogeneity underlying RNA modification-mediated regulatory mechanisms. The findings indicated that hypertension may affect RNA modifications and gene expression in aortic cells and may have an impact on AD. Further studies are suggested to elucidate the mechanism.
m6A is a type of dynamic and reversible RNA modification that plays critical roles in gene expression regulation29 and mRNA stability30 and homeostasis.31 RNA modification is involved in disease development and can be used in the clinic.32 However, the role of RNA modification in BP regulation is unknown. In the present study, we showed that searching for RNAm-SNPs in genomic loci was essential for a better understanding of GWAS signals. The enrichment of these RNAm-SNPs in protein-coding genes, particularly those involved in pathways related to pancreatic secretion, renin secretion, and immune responses, highlights the potential functional relevance of RNA modifications in physiological processes influencing BP. RNAm-SNPs in disease loci may be causal variants, as they can interrupt the modification and then disturb gene expression regulation.7 We successfully identified RNAm-SNPs in well-known pharmacologically active BP genes, including CYP11B1, PDE3B, HDAC7, ACE, SLC4A7, PDE1A, FRK, MTHFR, NPPA, CACNA1D, and HDAC9, emphasizing their potential role as therapeutic targets. The renin-angiotensin-aldosterone system (RAAS) is a well-described physiological system that is overactivated in hypertension. RNAm-SNPs in RAAS genes were identified and highlighted in our study. ACE, CREB1, PDE1A, PDE3B, GNAS, CACNA1D, and PLCB1 identified in this study are involved in the renin secretion pathway. In addition, the cell experiments showed that m6A methylation in BP genes (e.g., DDI2, MICB, UFL1, and FRK) may affect gene expression and that gene expression was associated with BP. This study showed that RNAm-SNPs may have impacts on the expression regulation of key BP genes. Therefore, RNA modification may play a role in BP regulation. Thus far, how RNAm-SNPs affect gene expression and how they contribute to BP regulation is unknown. The underlying mechanisms need to be clarified.
This study has some potential limitations. First, the identified RNAm-SNPs may represent only a subset of potential regulatory elements, and further functional validation is required to establish causality. Second, we did not consider interactions between RNA modifications and how environmental or lifestyle factors might interact with genetic and epigenetic factors to influence BP. Third, although we identified the BP-associated m6A-SNP, other RNA modification types were less found in this study, because data on other types of RNA modification were still scarce. Finally, the focus on aortic tissue provides insights into vascular smooth muscle cells, but exploring the broader implications in other cell types and tissues is warranted. Future studies should delve into the dynamic nature of RNA modifications, considering their temporal and spatial variations, to enhance our understanding of their regulatory roles.
In conclusion, our study unveils a complex interplay between genetic variants, RNA modifications, gene expression, and BP regulation. The identified RNAm-SNPs, particularly those associated with m6A modifications, present promising avenues for therapeutic interventions in hypertension. The associations with aortic structure and AD further emphasize the broader implications of RNA modifications in cardiovascular health. The findings indicated that hypertension may promote AD through the influence of RNA modifications and gene expression in aortic cells. As the field of RNA epigenetics continues to evolve, further investigations into the functional consequences of RNA modifications will undoubtedly enhance our understanding of cardiovascular diseases and pave the way for innovative therapeutic strategies.
AUTHOR CONTRIBUTIONS
Huan Zhang, Yuxi Chen, and Peng Xu wrote the first draft of the manuscript and revised the manuscript based on the authors' suggestions. Guideline panel members Dan Liu, Naqiong Wu, Laiyuan Wang, and Xingbo Mo critically reviewed the manuscript and provided additional information. Laiyuan Wang and Xingbo Mo were the chair of the panel and led the panel meeting. All authors approved the content.
ACKNOWLEDGMENTS
The study was supported by the National Natural Science Foundation of China (82173597, 82170480, 82073636, 81773508), the CAMS Innovation Fund for Medical Sciences (CIFMS) (2021-I2M-1-008), the Startup Fund from Soochow University (Q413900313, Q413900412), and a Project of the Priority Academic Program Development of Jiangsu Higher Education Institutions.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
The current study was approved by the Ethics Committee of Soochow University (ECSU-201800051).
Open Research
DATA AVAILABILITY STATEMENT
For original data, please contact the corresponding author.




