Utility of a Large Series of B-Cell Precursor Acute Lymphoblastic Leukemia Cell Lines as a Model System
Funding: This work was supported by JSPS KAKENHI Grant Number 23K24298 and Japan Research Foundation for Clinical Pharmacology.
ABSTRACT
Background
In B-cell precursor acute lymphoblastic leukemia (BCP-ALL), chromosomal translocations are strongly associated with prognoses. RNA sequencing (RNA-seq) is a powerful technology that reveals a close correlation between types of translocation and patterns of gene expression in clinical samples of BCP-ALL. Cancer cell lines are powerful research tools, and thus, we built a larger series of BCP-ALL cell lines and performed RNA-seq analysis to confirm their utility as a model system.
Methods
We performed RNA-seq in a total of 94 BCP-ALL cell lines, including 80 cell lines with 8 representative types of translocations.
Results
In the UMAP visualization, a close association was confirmed between the types of fusion genes and patterns of gene expression. In the cluster analysis of the gene expression profile, each type of fusion gene showed a clear association with the expression profile in the top 51 variable genes. Of clinical importance, the majority of the top variable genes in the BCP-ALL cell lines also showed a significant association with the types of fusion genes in the clinical samples. When an association of 125 cell cycle-related genes with the percentage of S and G2/M phases in 67 cell lines was evaluated, a significant positive correlation with cell cycle progression was confirmed in 10 cell cycle-related genes (HDAC2, CDC23, YWHAG, MAD2L1, CCNH, ANAPC7, CDC6, ANAPC5, ORC3, andRBX1). Moreover, significant upregulation and downregulation of 40 and 10 genes, respectively, were observed in the cell lines established at relapse compared with those established at diagnosis. Four (SP6, CCNE1, HIST1H2BH, and DECR2) and two (EVI2B and SYN1) of these genes were also significantly higher and lower, respectively, in the clinical samples at relapse than in those at diagnosis.
Conclusion
Large series of BCP-ALL cell lines is a powerful research tool for studying the mechanisms of leukemogenesis and the disease progression of BCP-ALL.
1 Introduction
In acute lymphoblastic leukemia (ALL), particularly in B-cell precursor ALL (BCP-ALL), chromosomal translocations, which are the most critical cytogenetic abnormalities, are highly associated with prognoses. Based on recent advances in next-generation sequencing technologies, many studies using clinical samples have demonstrated the utility of RNA sequencing (RNA-seq) in the molecular taxonomy of ALL [1-4]. RNA-seq is a powerful technology that enables us to simultaneously evaluate gene rearrangements and gene expression profiles in a single assay. As a result, RNA-seq reveals a close correlation between types of gene rearrangement and patterns of gene expression in BCP-ALL [1-4].
Cancer cell lines are powerful research tools for a better understanding of the biology and therapeutic responses in cancers. In particular, a large series of cancer cell lines is an outstanding platform for studying genomic diversity and identifying novel therapeutic targets in human cancers. The Cancer Cell Line Encyclopedia (CCLE) systematically profiles more than 1000 cell lines from diverse lineages, and the Dependency Map (DepMap) portal derived from CCLE is the most widely used genetic database for many cancers [5, 6].
As indicated in the CCLE studies [5, 6], leukemic cell lines are also useful research tools. However, the BCP-ALL cell lines available on the CCLE platform are currently limited to 36 cell lines. Despite the diverse genomic information for each cell line, this number might not be sufficient to evaluate a possible correlation between types of gene rearrangement and patterns of gene expression. Thus, we sought to build a larger series of BCP-ALL cell lines as a model system to understand the mechanisms of leukemogenesis and drug resistance. Since most BCP-ALL cell lines harbor chromosomal translocations, their gene expression profiles could show a significant association with their types of fusion genes. Thus, in the present study, we performed RNA-seq analysis in our large series of BCP-ALL cell lines and confirmed a close correlation between types of fusion genes and gene expression profiles. We further evaluated the association of the gene expression profile with cell proliferation and disease progression.
2 Materials and Methods
2.1 Cell Lines
We used 94 BCP-ALL cell lines (Table 1), including 21 MEF2D-rearranged (MEF2D-R), 17 BCR::ABL1, 14 KMT2A-R, 14 TCF3::PBX1, 5 ETV6::RUNX1, 4 TCF3::HLF, 3 Ph-like, and 2 DUX4-R cell lines. All cell lines were maintained in RPMI1640 medium supplemented with 10% fetal calf serum (FCS) in a humidified atmosphere of 5% CO2 at 37°C. Forty-four cell lines had been sequentially established in our laboratory since 1978. In brief, heparinized sterile mononuclear cells isolated from peripheral blood or bone marrow by Ficoll density gradient centrifugation were cultured in RPMI1640 medium supplemented with 20% FCS until they proliferated exponentially. Research protocol, including written informed consent, was approved by the University of Yamanashi, Certified Review Board: Approval No. 1231.
| Cell lines | Fusion gene | Establishment | S-G2/M (%) | Calalog no. | CCLE |
|---|---|---|---|---|---|
| 697 | TCF3::PBX1 | At relapse | NA | DSMZ ACC 42 | ○ |
| Kasumi2e | TCF3::PBX1 | At relapse | 38 | JCRB1395 | ○ |
| KOPN34a | TCF3::PBX1 | At diagnosis | 43 | NA | NA |
| KOPN36a | TCF3::PBX1 | At relapse | 49 | NA | NA |
| KOPN54a | TCF3::PBX1 | At relapse | 32 | NA | NA |
| KOPN63a | TCF3::PBX1 | At relapse | 50 | NA | NA |
| PreALP | TCF3::PBX1 | At diagnosis | NA | NA | NA |
| RCH | TCF3::PBX1 | At relapse | NA | DSMZ ACC 548 | NA |
| SCMC-L1l | TCF3::PBX1 | NA | 37 | NA | NA |
| THP4c | TCF3::PBX1 | At diagnosis | 28 | NA | NA |
| YAMN90Ra | TCF3::PBX1 | At relapse | 34 | NA | NA |
| YAMN92a | TCF3::PBX1 | At relapse | 29 | NA | NA |
| YcuB6b | TCF3::PBX1 | At diagnosis | 28 | DSMZ ACC 966 | NA |
| YcuB8b | TCF3::PBX1 | At diagnosis | 33 | NA | NA |
| KOPN68a | ETV6::RUNX1 | At relapse | 30 | NA | NA |
| KOPN79a | ETV6::RUNX1 | At relapse | 16 | NA | NA |
| KOPN87a | ETV6::RUNX1 | At relapse | 32 | NA | NA |
| MBITd | ETV6::RUNX1 | At diagnosis | NA | NA | NA |
| Reh | ETV6::RUNX1 | At relapse | NA | ATCC CRL-8286 | ○ |
| KOCL33a | KMT2A-R (KMT2A::MLLT1) | At diagnosis | 51 | NA | NA |
| KOCL44a | KMT2A-R (KMT2A::MLLT1) | At relapse | 34 | NA | NA |
| KOCL45a | KMT2A-R (KMT2A::AFF1) | At relapse | 56 | NA | NA |
| KOCL50a | KMT2A-R (KMT2A::AFF1) | At relapse | 57 | NA | NA |
| KOCL51a | KMT2A-R (KMT2A::MLLT1) | At diagnosis | 35 | NA | NA |
| KOCL58a | KMT2A-R (KMT2A::AFF1) | At diagnosis | 38 | NA | NA |
| KOCL69a | KMT2A-R (KMT2A::AFF1) | At relapse | 43 | NA | NA |
| KOCL77a | KMT2A-R (KMT2A::AFF1) | At relapse | NA | NA | NA |
| KOPB26a | KMT2A-R (KMT2A::MLLT3) | At relapse | 57 | NA | NA |
| KOPN1a | KMT2A-R (KMT2A::MLLT1) | At relapse | 41 | NA | NA |
| KOPN35a | KMT2A-R (KMT2A::MLLT10) | At relapse | 48 | NA | NA |
| RS411 | KMT2A-R (KMT2A::AFF1) | At relapse | NA | ATCC CRL-1873 | ○ |
| THP8c | KMT2A-R (KMT2A::AFF1) | At diagnosis | 32 | NA | NA |
| YACL95 | KMT2A-R (KMT2A::MLLT3) | At diagnosis | 40 | NA | NA |
| Kasumi8e | BCR::ABL1 | At relapse | NA | JCRB 1403 | NA |
| KCB1b | BCR::ABL1 | At diagnosis | 11 | NA | NA |
| KOPN30bia | BCR::ABL1 | At relapse | 23 | NA | NA |
| KOPN55bia | BCR::ABL1 | At relapse | 15 | NA | NA |
| KOPN56a | BCR::ABL1 | At relapse | 36 | NA | NA |
| KOPN57bia | BCR::ABL1 | At diagnosis | 34 | NA | NA |
| KOPN66bia | BCR::ABL1 | At relapse | 33 | NA | NA |
| KOPN83bia | BCR::ABL1 | At relapse | 24 | NA | NA |
| Nalm1 | BCR::ABL1 | NA | NA | ATCC CRL-1567 | NA |
| Nalm27 | BCR::ABL1 | At diagnosis | 37 | JCRB 1830 | NA |
| PALL-2 | BCR::ABL1 | At relapse | NA | JCRB 1345 | NA |
| SK9j | BCR::ABL1 | At relapse | 38 | NA | NA |
| SU-Ph2g | BCR::ABL1 | At relapse | 30 | NA | NA |
| TCCYh | BCR::ABL1 | NA | 40 | NA | NA |
| TMD5 | BCR::ABL1 | At diagnosis | NA | JCRB IFO50516 | NA |
| YAMN73a | BCR::ABL1 | At relapse | 32 | NA | NA |
| YAMN91a | BCR::ABL1 | At diagnosis | 34 | NA | NA |
| HBL3f | MEF2D-R (MEF2D::BCL9) | At relapse | NA | NA | NA |
| Kasumi7e | MEF2D-R (MEF2D::HNRNPUL1) | At relapse | NA | JCRB 1401 | NA |
| Kasumi9e | MEF2D-R (MEF2D::HNRNPUL1) | At relapse | NA | JCRB 1409 | NA |
| KOPN39a | MEF2D-R (MEF2D::BCL9) | At diagnosis | 44 | NA | NA |
| KOPN46a | MEF2D-R (MEF2D::HNRNPUL1) | At diagnosis | NA | NA | NA |
| KOPN61a | MEF2D-R (MEF2D::BCL9) | At diagnosis | 36 | NA | NA |
| KOPN62a | MEF2D-R (MEF2D::BCL9) | NA | 38 | NA | NA |
| KOPN70a | MEF2D-R (MEF2D::BCL9) | At relapse | 39 | NA | NA |
| KOPN71a | MEF2D-R (MEF2D::BCL9) | At diagnosis | 42 | NA | NA |
| KOS20 | MEF2D-R (MEF2D::BCL9) | At relapse | 45 | NA | NA |
| L-ASKc | MEF2D-R (MEF2D::BCL9) | At diagnosis | 36 | NA | NA |
| L-KUMc | MEF2D-R (MEF2D::DAZAP1) | At relapse | 40 | NA | NA |
| LC4-1 | MEF2D-R (MEF2D::HNRNPUL1) | NA | NA | JCRB 0114 | ○ |
| MBMYd | MEF2D-R (MEF2D::HNRNPUL1) | At diagnosis | 46 | NA | NA |
| P30/OHK | MEF2D-R (MEF2D::HNRNPUL1) | At relapse | 43 | RCB 1938 | ○ |
| THP5c | MEF2D-R (MEF2D::HNRNPUL1) | At diagnosis | 44 | NA | NA |
| THP7c | MEF2D-R (MEF2D::BCL9) | At diagnosis | 52 | NA | NA |
| YAMN74a | MEF2D-R (MEF2D::BCL9) | At relapse | 41 | NA | NA |
| YAMN96a | MEF2D-R (MEF2D::HNRNPUL1) | At diagnosis | NA | NA | NA |
| YcuB4b | MEF2D-R (MEF2D::BCL9) | At diagnosis | 41 | DSMZ ACC 962 | NA |
| YcuB7b | MEF2D-R (MEF2D::BCL9) | At relapse | 33 | NA | NA |
| Endokunk | TCF3::HLF | At relapse | 39 | NA | NA |
| HAL-O1i | TCF3::HLF | At relapse | 31 | RCB 0540 | ○ |
| UOC-B1 | TCF3::HLF | At relapse | NA | NA | NA |
| YcuB2b | TCF3::HLF | At relapse | 46 | DSMZ ACC 961 | NA |
| KOPN49a | IgH::CRLF2 | At relapse | 24 | NA | NA |
| NAGL-1 | IgH::CRLF2 | NA | NA | JCRB IFO50479 | NA |
| YcuB5b | P2RY8::CRLF2 | At diagnosis | 19 | DSMZ ACC 964 | NA |
| KOPN84a | DUX4-R | At relapse | 43 | NA | NA |
| Nalm6 | DUX4-R | At relapse | NA | ATCC CRL-3273 | ○ |
| CCRF-SB | Others | NA | NA | JCRB 0032 | NA |
| KCB2b | Others | At relapse | NA | NA | NA |
| KCB4b | Others | At relapse | 21 | NA | NA |
| KCB6b | Others | NA | NA | NA | NA |
| KCB7b | Others | At diagnosis | NA | NA | NA |
| KOPB38a | Others | NA | NA | NA | NA |
| KOPB59a | Others | NA | NA | NA | NA |
| KOPN32a | Others | At relapse | 41 | NA | NA |
| KOPN40a | Others | At relapse | 34 | NA | NA |
| KOPN75a | Others | At diagnosis | 45 | NA | NA |
| KOPN85a | Others | At relapse | 28 | NA | NA |
| MBKGd | Others | At relapse | 54 | NA | NA |
| MBOKd | Others | At relapse | 48 | NA | NA |
| SCMC-L2l | Others | NA | 49 | NA | NA |
- Abbreviations: ATCC, American Type Culture Collection; DSMZ, German Collection of Microorganisms and Cell Cultures; JCRB, Japanese Collection of Research Bioresources; NA, not abailable; RCB, RIKEN Cell Bank.
- a In house.
- b Yokohama City University and Kanagawa Children's Medical Center (Dr. H. Goto).
- c Tohoku University (Dr. M. Minegishi).
- d Mie University Graduate School of Medicine (Dr. S. Iwamoto).
- e Hiroshima University (Dr. T. Inaba).
- f Fukushima Medical University (Dr. H. Hojo).
- g Kindai University Faculty of Medicine (Dr. Y. Maeda).
- h Tochigi Cancer Center (Dr. Y. Sato).
- i Dana-Farber Cancer Institute, Boston, MA (Dr. A. T. Look).
- j Tokyo Medical University (Dr. S. Okabe).
- k Iwate Medical University (Dr. M. Endo).
- l Saitama Children's Medical Center (Dr. J. Takita).
2.2 RNA-Seq Analysis
RNA-seq was performed by Rhelixa Co. Ltd. (Tokyo, Japan) using total RNA extracted with a RNeasy Plus Mini Kit (QIAGEN, Hilden, Germany). Libraries were prepared using a NEBNext Ultra II Directional RNA Library Prep Kit (New England BioLabs, Beverly, MA) and then sequenced on an Illumina NovaSeq 6000 with 150 × 2 paired-end reads. Reads were trimmed to remove adapters and low-quality bases with the TrimGalore! (v.0.6.6) graphical interface tool. The trimmed reads were aligned to the reference genome (hg38) using the RNA-seq aligner STAR (v.2.1.0) with RSEM (v.1.1.17), according to a previous report [7]. The total number of reads and gene lengths among the samples were corrected using transcripts per million (TPM). Differentially expressed genes between the two groups were analyzed using the R package DEseq2 (v.1.38.3). For two-dimensional UMAP, which was performed by using the R package umap (v.0.2.10.0), variable genes were determined by analyzing differentially expressed genes between the cell lines with and without each fusion gene. We carried out fusion gene detection using FusionCatcher (v1.33) with default parameters [8]. The analysis was performed on raw fastq file of each cell line, utilizing the human genome reference GRCh38, and a summary of matrix is presented in Table S1.
2.3 Cell Cycle Analysis
Each cell line was fixed with 70% ethanol 1 day after medium exchange, stained with propidium iodide (PI) (Sigma, St. Louis, MO), and analyzed by flow cytometry. The median percentages of the S and G2/M phases in three independent analyses were previously determined in 67 cell lines [9].
2.4 Statistical Analyses
Simple linear regression of gene expression and cell cycle was performed using GraphPad Prism (v.10.0.2) software. The Mann–Whitney U test was applied to compare the two groups.
3 Results
3.1 Association of Types of Fusion Gene With Gene Expression Profile in BCP-ALL Cell Lines
To confirm whether the types of fusion genes show an association with the gene expression profile in BCP-ALL cell lines, we focused on 80 cell lines whose fusion genes were previously determined by RNA capture sequencing analysis [9]. These cell lines harbor the following eight representative types of fusion genes: MEF2D-R, BCR::ABL1, KMT2A-R, TCF3::PBX1, ETV6::RUNX1, TCF3::HLF, Ph-like, and DUX4-R. First, RNA-seq identified the type of fusion gene in each cell line, which was the same as the one previously determined. Then, we performed UMAP analysis based on the 1554 most variable genes. Two-dimensional UMAP (Figure 1a) visualized that TCF3::PBX1 cell lines (yellow), MEF2D-R cell lines (dark red), KMT2A-R cell lines (red), and the other cell line types exclusively clustered with each other. Moreover, although relatively close to each other in visualization, ETV6::RUNX1 cell lines (moss green), DUX4-R cell lines (light blue), TCF3::HLF cell lines (pink), and BCR::ABL1 cell lines (blue) showed exclusive expression profiles. Of note, the gene expression profile in the three Ph-like cell lines [two IgH::CRLF2 (light green) and one P2RY8::CRLF2 (brown) cell lines] completely overlapped with those in the BCR::ABL1 (blue) cell lines, as previously observed in the clinical samples [3, 4].

Next, we applied the cluster analysis to further investigate an association of the gene expression profile with 8 representative types of fusion genes in 80 cell lines. As shown in the heatmap based on the cluster analysis (Figure 1b), each type of fusion gene showed a clear association with the expression profile in the top 51 variable genes. The specific higher expression of PBX1 in TCF3::PBX1 cell lines, ABL1 in BCR::ABL1 cell lines, HLF in TCF3::HLF cell lines, and CRLF2 in Ph-like cell lines is assumed to be derived from reads of the fusion genes. To confirm the significance of these genes in the clinical samples, we developed a gene expression heatmap of the same genes in 341 childhood BCP-ALL cases using the RNA-seq data in a public database of St. Jude Children's Research Hospital (https://pecan.stjude.cloud/). We evaluated 48 genes except for EBLN3P, HLF, and PAN3, which were specifically associated with Ph-like (CRLF2-R), TCF3::HLF, and KMT2A-R cell lines, respectively, due to lack of expression data. As shown in Figure S1, the majority of these genes showed a significant association with the types of fusion genes, particularly in TCF3::PBX1, ETV6::RUNX1, KMT2A-R, and MEF2D-R samples. Subsequently, we performed UMAP analysis of cell lines and clinical samples using these 48 variable genes (Figure 1c) and confirmed almost the same patterns of association between cell lines and clinical samples. These findings in our series of BCP-ALL cell lines reveal a close association between the types of fusion genes and patterns of gene expression, consistent with the findings in the clinical samples [3, 4].
3.2 Association of Cell Cycle-Related Genes With Cell Cycle Progression in BCP-ALL Cell Lines
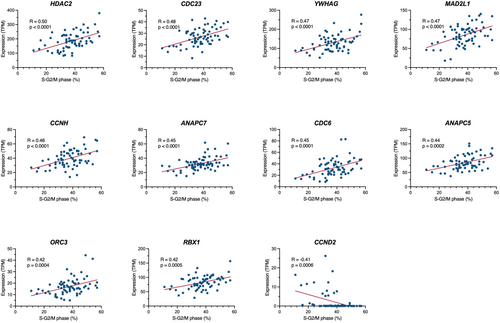
Leukemic cell lines generally show proliferative potential in vitro, which may at least partly be associated with the proliferative potential of leukemia cells in vivo. Thus, we applied the RNA-seq data to identify the cell cycle-related genes that are involved in the cell cycle progression of BCP-ALL cell lines. We evaluated the association of 125 cell cycle-related genes annotated in the KEGG pathway database (hsa04110) with the percentage of S and G2/M phases in 67 BCP-ALL cell lines, in which cell cycle analysis was previously performed by PI staining. We found a significant positive correlation (r > 0.4) in 10 genes (Figure 2), including HDAC2, CDC23, YWHAG, MAD2L1, CCNH, ANAPC7, CDC6, ANAPC5, ORC3, and RBX1. In contrast, a significant negative correlation (r < −0.4) was unexpectedly observed in CCND2, which encodes cyclin D2 (Figure 2). These observations suggest that at least some of these genes might be involved in the cell cycle progression of BCP-ALL cell lines.
3.3 Identification of Genes Associated With Disease Progression in BCP-ALL
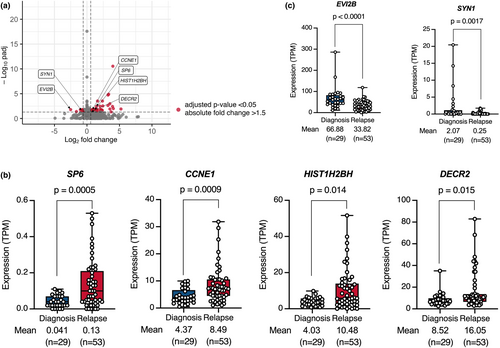
Because the relapsed cases had poor prognoses, identification of the gene expression profile associated with relapse is important for improved therapeutic outcomes. Using 82 BCP-ALL cell lines with establishment information, we compared gene expression profiles between cell lines established at diagnosis (n = 29) and those established at relapse (n = 53). As visualized in a volcano plot (Figure 3a), we identified 40 and 10 genes that were significantly upregulated and downregulated, respectively, in the cell lines established at relapse. Then, we compared the expression levels of these 50 genes between the clinical samples at diagnosis and those at relapse using a public database at St. Jude Children's Research Hospital. Of note, as observed in our BCP-ALL cell lines (Figure 3b,c), four (SP6, CCNE1, HIST1H2BH, and DECR2) and two (EVI2B and SYN1) genes were significantly higher and lower in the samples at relapse than in those at diagnosis, respectively (Figure S2). Moreover, similar patterns of difference were separately observed in the cell lines with four representative types of fusion genes (Figure S3). Accordingly, at least some of these genes might be involved in disease progression.
4 Discussion
In the present study, RNA-seq analysis was performed in a large number of BCP-ALL cell lines. In the two-dimensional UMAP analysis, 80 cell lines harboring 8 representative fusion genes were exclusively clustered with each other. In particular, we observed that the gene expression profiles of three Ph-like cell lines, including two IgH::CRLF2 and one P2RY8::CRLF2 cell line, overlapped those of BCR::ABL1 cell lines, as previously demonstrated in the clinical samples [3, 4]. Moreover, in the cluster analysis of the gene expression profile of 80 cell lines with 8 representative fusion genes, each type of fusion gene showed a clear association with the expression profile in the top 51 variable genes.
In the present study, by using the FusionCatcher, we failed to detect fusion genes in four cell lines, in which major types of fusion transcripts were already detectable by the RT-PCR analysis. Thus, simultaneous application of other fusion detectors such as the STAR-Fusion might be helpful to improve sensitivity. Among the 14 cell lines without 8 major types of fusion genes, no specific fusion genes were found in 11 cell lines, while the significance of the identified fusion gene in the remaining 3 cell lines is currently under investigation.
Of note, among these top variable genes, WNT16 [10], SLC51A [11], MEIS1 [12], and PTPRZ1 [13] were reportedly specifically overexpressed in TCF3::PBX1, ETV6::RUNX1, KMT2A-R, and MEF2D-R clinical samples, respectively. Of clinical importance, the majority of these top variable genes in BCP-ALL cell lines also showed a significant association with the types of fusion genes in the clinical samples when applied to the RNA-seq data in a public database of 341 childhood BCP-ALL cases at St. Jude Children's Research Hospital. To the best of our knowledge, although it has already been confirmed in clinical samples [3, 4], this is the first study confirming a close association between types of fusion genes and patterns of gene expression in a large series of BCP-ALL cell lines. Additionally, we re-investigated the association of fusion gene types with copy number alterations (Figure S4), which were previously evaluated by multiplex ligation-dependent probe amplification analysis in the majority of cell lines [14]. As investigated in previous studies of clinical samples [15, 16], we confirmed the exclusive association of IKZF1 deletion in BCR::ABL1 cell lines and fewer copy number alterations in KMT2A-R cell lines. These observations suggest similarities in the patterns of copy number alteration between BCP-ALL cell lines and clinical samples. Accordingly, our observations indicate that BCP-ALL cell lines are useful tools to investigate the biological significance of types of fusion genes, at least in terms of gene expression profiles.
We next applied the RNA-seq data to identify the cell cycle-related genes that are involved in the cell cycle progression of BCP-ALL cell lines and identified 10 genes, including HDAC2, CDC23, YWHAG, MAD2L1, CCNH, ANAPC7, CDC6, ANAPC5, ORC3, and RBX1, which were positively correlated with cell cycle progression. Among these 10 genes, our literature search revealed the involvement of the following 6 genes in the cell cycle progression of cancers.
HDAC2, a histone deacetylase, reportedly promoted cell cycle progression of breast cancer cell lines, since CRISPR/Cas9-mediated knockout induced reduction of the cells in S and G2/M phases [17]. CDC23, one of the APC subunits involving cell mitosis, was reportedly associated with cell cycle progression in liver cancer cell lines, since shRNA-mediated knockdown repressed cell proliferation [18]. YWHAG, one of the 14-3-3 phospho-serine/phospho-threonine binding protein lines, promoted cell proliferation in the pancreas cancer cell lines, since expression vector-mediated overexpression enhanced cell growth [19].
MAD2L1, a component of the mitotic spindle assembly checkpoint, was reportedly involved in cell cycle progression of colon cancer cell lines, since siRNA-mediated knockdown induced a reduction of the cells in the S and G2/M phases [20]. CDC6, a DNA replication initiation factor, reportedly promoted cell cycle progression, since siRNA-mediated knockdown blocked the G1/S transition in the HeLa cell line [21]. RBX1, an E3 ubiquitin ligase activator, reportedly promoted cell cycle progression in melanoma and multiple myeloma cell lines, since lentiviral vector-mediated overexpression enhanced progression into the S phase [22].
Although no previous publications regarding involvement in tumorigenesis exist, the other four genes (CCNH, ANAPC7, ANAPC5, and ORC3) seem to be functionally consistent with cell cycle progression. On the other hand, we identified that the CCND2 gene had a significant negative correlation with cell cycle progression in BCP-ALL cell lines. However, 11 out of 15 cell lines with higher CCDN2 expression (TPM value > 5) were BCR::ABL1 cell lines, in which cyclin D2 is reportedly upregulated by the tyrosine kinase activity of the BCR::ABL1 protein [23, 24]. Thus, a negative correlation between cell cycle progression and CCND2 might be simply due to a relatively slower cell cycle progression in BCR::ABL1 cell lines. Taken together, at least some of these genes might be involved in the cell cycle progression of BCP-ALL cell lines and, subsequently, BCP-ALL cells in vivo, although further direct evaluation is required.
Finally, we compared gene expression profiles between cell lines established at diagnosis and those established at relapse and identified 40 and 10 genes that were significantly upregulated and downregulated, respectively, in the cell lines established at relapse. As a potential limitation of this analysis, clonal selection could occur during establishment in the cell lines established at diagnosis. Among those genes, we confirmed that 4 genes, including SP6, CCNE1, HIST1H2BH, and DECR2, and 2 genes, including EVI2B and SYN1, were significantly higher and lower in the clinical samples at relapse than in those at diagnosis, respectively. Of interest, our literature search revealed the possible involvement of all four upregulated and one of two downregulated genes in tumor progression. SP6, a Krüppel-like family transcription factor, was reportedly involved in cell proliferation in dental pulp mesenchyme by enhancing canonical Wnt/β-catenin signaling, since expression vector-mediated overexpression induced cellular accumulation of β-catenin [25]. CCNE1, a member of the cyclin family, was reportedly involved in cell cycle progression in breast cancer patients, since higher expression was associated with clinical resistance to a CDK4/6 inhibitor [26]. HIST1H2BH, a member of the H2B histone family, was reportedly involved in cell proliferation in multiple myeloma cell lines, since siRNA-mediated knockdown inhibited proliferation [27]. DECR2, a 2,4-dienoyl-CoA reductase 2, was reportedly involved in cell proliferation in prostate cancer cell lines, since lentiviral vector-mediated overexpression intensified cell growth [28].
Conversely, EVI2B, a single-pass type I transmembrane glycoprotein, was reportedly involved in tumor immunity, since the cytotoxic activity of CD8+ T cells was intensified in melanoma patients with its higher expression [29]. Taken together, at least some of these genes might be involved in the disease progression of BCP-ALL and, subsequently, poor outcomes in the relapsed cases, although further evaluation is required.
5 Conclusion
In the present study, using RNA-seq analysis of large series of BCP-ALL cell lines, we first demonstrated a close association between types of fusion genes and patterns of gene expression. We also identified a couple of cell cycle-related genes that were positively correlated with cell cycle progression and relapsed-related genes that were associated with disease relapse in the clinical samples. Although full multi-omics analyses would be more informative, these observations revealed that our large series of BCP-ALL cell lines is a powerful research tool for studying the mechanisms of leukemogenesis and the disease progression of BCP-ALL.
Author Contributions
Minori Tamai: conceptualization (lead), data curation (lead), formal analysis (lead), funding acquisition (lead), investigation (lead), methodology (lead), software (lead), validation (lead), visualization (lead), writing – original draft (lead). Chiaki Komatsu: formal analysis (supporting), investigation (supporting). Keiko Kagami: formal analysis (supporting), investigation (supporting). Shin Kasai: data curation (supporting). Koshi Akahane: data curation (supporting). Kumiko Goi: data curation (supporting). Kanji Sugita: data curation (supporting), resources (lead). Chihiro Tomoyasu: data curation (supporting), investigation (supporting), methodology (supporting). Toshihiko Imamura: data curation (supporting), investigation (supporting), methodology (supporting). Hiroaki Goto: data curation (supporting), resources (lead). Takeshi Inukai: conceptualization (supporting), data curation (supporting), funding acquisition (lead), investigation (supporting), methodology (supporting), project administration (lead), resources (lead), supervision (lead), validation (supporting), visualization (supporting), writing – review and editing (lead).
Ethics Statement
Approval of the research protocol, including written informed consent, by the University of Yamanashi, Certified Review Board: Approval No. 1231 (2018 July 9th).
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The RNA-seq data of 80 BCP-ALL cell lines with 8 major fusion genes are available under the DDBJ BioProject database: PRJDB18849.