Utilization of cytologic cell blocks for targeted sequencing of solid tumors
Abstract
Background
Targeted sequencing of cytologic samples has significantly increased in recent years. With increasing numbers of clinical trials for variant specific therapeutics, validating a comprehensive assay for cytologic samples has become clinically important.
Aim
For this study, a retrospective review of cytologic cell blocks from fine needle aspirations and fluid specimens was performed.
Methods
Two hundred twenty six total cases of solid tumor malignancies were identified, of which 120 cases and 20 lymph node negative controls were sequenced for the Oncomine Comprehensive Assay. Cytology and surgical specimen correlation was performed in a subset of cases. Statistical analysis to determine variant concordance was performed.
Results
Within the 117 cases sequenced, a total of 347 pathogenic variants were detected. Of the 117 cases, 32 cases (27.4%) would qualify for FDA approved targeted therapy according to the current guidelines, and an additional 23 cases (19.7%) would qualify for clinical trial based on pathogenic variants detected.
Discussion
With over 27% of cases in our cohort qualifying for some form of targeted therapy, our study shows the importance of providing comprehensive molecular diagnostic options. Despite only half of the cytology cases in the review period having enough material to be sequenced, overall approximately 27% of patients in this cohort would have benefitted from this service.
1 INTRODUCTION
Utilization of cytologic samples for clinical testing in the molecular pathology laboratory has greatly increased over recent years. As next-generation sequencing (NGS) technologies have become dominant in the diagnostic and therapeutic arenas, it is essential to optimize assays for which low input of DNA can be used to sequence large numbers of clinically actionable targets in a highly accurate and reproducible manner. This will permit comprehensive overviews of the driver mutations for targeted therapy even in very small samples of tumor. Several publications have addressed the use of cytologic smear preparations and the minimum amount of tissue needed for adequate DNA/RNA extraction from a cytology sample.1-3 More recent studies have shown the potential of formalin-fixed and paraffin-embedded (FFPE) cytology cell blocks as an additional source of molecular testing material without the destruction of diagnostic cytologic smears.4, 5 In addition, there are preanalytic variables that are specific to cytology samples.1 With a small amount of tissue as seen in cytologic samples, variables such as tumor heterogeneity and low percentage of tumor cells may lead to false-negative results. However, in cases of metastasis at the time of presentation or in cases where a fine needle aspiration (FNA) is the only procedure the patient can tolerate for sample collection, utilization of these small samples is invaluable for patient care.
For utilizing small samples, targeted panels are an ideal way to assess a tumor for clinically actionable mutations compared to PCR and Sanger sequencing methods. With many FDA-approved drugs available for various types of solid tumors harboring mutations in EGFR, BRAF, BRCA, NTRK, PIK3CA, etc., sequencing these targets has never been more important for patient care. At our institution, a medium-sized university hospital, we have elected to use a comprehensive solid tumor panel that targets important driver genes for therapy, tumor progression, and prognosis across all types of solid tumors. To our knowledge, this is the first study utilizing cytology cell blocks across multiple cancer types for comprehensive targeted sequencing for solid tumors to simulate routine diagnostic molecular oncology workflow. In the present study, we reviewed cytology cell blocks from 2013–2017 to identify cases with remaining malignant cells to be used for targeted panel sequencing. In order to test the viability of cytologic cell block samples as an alternative to surgical pathology material, archival cytology cell blocks from cytology fluid specimens and FNA material was utilized to simulate samples in a standard practice workflow.
2 MATERIALS AND METHODS
2.1 Case selection
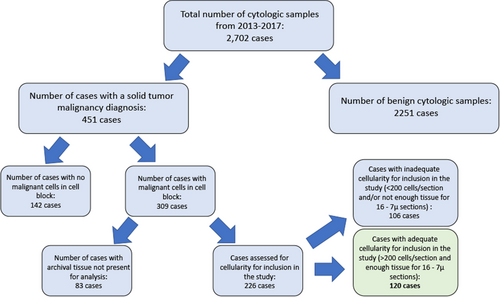
Institutional Review Board approval for this study was obtained from the University of Illinois at Chicago. A case retrieval search was performed at the University of Illinois at Chicago pathology archives for all FNAs and cytology fluid samples that had a corresponding cytology cell block from August 2013 through May 2017. FNA specimens and fluid samples were chosen as all of these cases reflexively have a cell block performed. All samples were initially fixed in CytoLyt and transferred in pellet form into formalin for a minimum of 6 h prior to being placed on the processor in the histology laboratory. A total of 2702 cases were identified during this time period. The diagnoses of these cases were reviewed to identify all cases that had a malignant solid tumor neoplasm (this included carcinomas and soft tissue tumors). A total of 451 cases were identified as being positive for a solid tumor malignancy, and the cell block H&E slides for these cases were reviewed to confirm presence of malignant cells. Approximately 30% of these cases were found to have no malignant cells within the cell block sections and were confirmed to have had the diagnosis of malignancy rendered on the cytospin or FNA smear preparations; these cases were excluded from the study. Another 20% of cases identified were excluded from the study due to cell blocks or slides missing from the archives.
Of the remaining 226 cases, 106 (47%) cases were found to have inadequate numbers of tumor cells on cell block H&E sections (cases with less than 200 tumor cells per H&E section were considered inadequate and not included in the study) or inadequate tissue remaining on the paraffin block for 16 unstained slides to be cut for the molecular analysis performed in this study. A total of 120 cases were identified with enough archival tissue remaining to proceed with molecular analysis (Figure 1).
In addition to the cytology cell blocks for 120 cases, we reviewed the pathology archival material for any surgical pathology case for the patient cohort that had the same diagnosis as the cytology sample. Many of our cases within the cohort were cytologic fluid samples that represented metastatic disease where surgical resection was not an option (Table 1). In addition, our center is a tertiary care referral center and many smaller community practices send patients to our clinics for endoscopic diagnostic procedures, and therefore, resection specimens are not present in our archives for comparison in many lung and gastrointestinal malignancies. Due to these conditions, we were able to find surgical samples for only 18 cases that corresponded to the cytology sample and were included in the study for comparison of NGS results.
| Case ID | Diagnosis | Age @ diagnosis | Gender | Tissue Type | Clinical/Pathologic Stage at Diagnosis | Clinical Hx (Hx) | Primary/ Metastasis |
|---|---|---|---|---|---|---|---|
| Case 1 | Lung Adenocarcinoma | 69 | F | Pleural fluid, left | Stage IVB; pT4pM1c | Contralateral lung, bone, and brain metastasis at the time of diagnosis | Met |
| Case 2 | Lung Adenocarcinoma | 58 | F | FNA, right hilar mass | pT2apN0 | — | Recurrence |
| Case 3 | Lung Adenocarcinoma | 73 | F | Pericardial fluid | Stage IVA; pT4pN3pM1a | Pleural fluid and pericardial fluid positive for malignancy at the time of diagnosis | Met |
| Case 4 | Lung Adenocarcinoma | 64 | F | Pleural fluid | Stage IVB | Supraclavicular lymph node and brain metastasis at the time of diagnosis | Met |
| Case 5 | Lung Adenocarcinoma | 54 | F | Pleural fluid | Stage IV | Spine metastasis at the time of diagnosis | Met |
| Case 6 | Lung Adenocarcinoma | 66 | M | Pericardial fluid | Stage IIIA | Pericardial malignant effusion and positive lymph nodes at the time of diagnosis | Met |
| Case 7 | Lung Adenocarcinoma | 58 | F | Pleural fluid | Stage IV | Bone metastasis at the time of diagnosis | Met |
| Case 8 | Lung Adenocarcinoma | 52 | F | FNA, lymph node | at least Stage IIIB | — | Met |
| Case 9 | Lung Adenocarcinoma | 37 | M | Pleural fluid | — | Malignant pleural effusion at the time of diagnosis | Met |
| Case 10 | Lung Adenocarcinoma | 80 | F | Pleural fluid | — | — | Met |
| Case 11 | Lung adenocarcinoma | 37 | F | Pericardial fluid | Stage IVB | — | Met |
| Case 12 | Lung Adenocarcinoma | 71 | F | TBNA-EBUS guided | Stage IV | — | Primary |
| Case 13 | Squamous cell carcinoma | 62 | F | TBNA-EBUS guided | Stage IIIB | — | Primary |
| Case 14 | Lung Adenocarcinoma | 60 | F | TBNA-EBUS guided | Stage IVA | Brain metastasis at the time of diagnosis | Primary |
| Case 15 | Lung Adenocarcinoma | 72 | F | Pleural fluid | Stage IV | — | Met |
| Case 16 | Lung Adenocarcinoma | 73 | M | TBNA-EBUS guided, lung mass | — | — | Primary |
| Case 17 | Lung Adenocarcinoma | 55 | F | TBNA-EBUS guided, right bronchus intermedius mass | Stage IVB; pT2b pN3 pM1c | — | Primary |
| Case 18 | Lung Adenocarcinoma | 69 | F | TBNA-EBUS guided, right lower lobe lung mass | Stage IIIB, pT2b pN3 | Original tumor diagnosed 7 years before presentation at an outside institution | Recurrence |
| Case 19 | Lung Adenocarcinoma | 61 | F | TBNA-EBUS guided | Stage IV | Metastasis known at the time of presentation | Primary |
| Case 20 | Lung Adenocarcinoma | 63 | F | TBNA-EBUS guided, mediastinal mass | Stage I, pT1 pN0 | Original tumor 15 years prior to presentation | Recurrence |
| Case 21 | Lung Adenocarcinoma | 71 | M | Pleural fluid | Stage IVB | Malignant pleural effusion at diagnosis | Met |
| Case 22 | Lung Adenocarcinoma | 65 | M | Pleural fluid | at least Stage III | — | Met |
| Case 23 | Lung adenocarcinoma | 66 | M | TBNA-EBUS guided, left lower lobe lung mass | Stage IIIA | — | Primary |
| Case 24 | Lung adenocarcinoma | 68 | M | TBNA-EBUS guided, right hilar mass | Stage IV | Brain metastasis at the time of diagnosis | Primary |
| Case 25 | Lung adenocarcinoma | 50 | M | Pleural fluid | Stage IVB | Malignant pleural effusion at the time of diagnosis | Recurrence |
| Case 26 | Lung adenocarcinoma | 75 | M | TBNA-EBUS guided, lymph node | Stage IV | — | Met |
| Case 27 | Lung SqCC | 61 | M | TBNA-EBUS guided, hilar mass | Stage IIIB | Hx of TB | Recurrence |
| Case 28 | Lung SqCC | 65 | F | TBNA-EBUS guided, lymph node | Stage IVA | Hx of LUL lung mass | Met |
| Case 29 | Lung SqCC | 64 | M | TBNA-EBUS guided, lymph node | At least Stage IIIB | Hx of right lower lobe lung mass | Met |
| Case 30 | Lung SqCC | 72 | F | TBNA-EBUS guided, mediastinal lymph node | Stage IIIB | Hx of smoking with lung mass | Met |
| Case 31 | Lung SqCC | 69 | M | TBNA-EBUS guided, right lung mass | Stage IV | — | Primary |
| Case 32 | Lung SqCC | 55 | M | TBNA-EBUS guided, lung mass | Stage IIIA | — | Primary |
| Case 33 | Lung SqCC | 62 | M | TBNA-EBUS guided, RUL mass | At least Stage IIIA | — | Primary |
| Case 34 | Lung SqCC | 82 | F | TBNA-EBUS guided, left lung mass | At least Stage III | — | Primary |
| Case 35 | Small cell carcinoma, Lung | 59 | M | Pleural fluid | Stage IV | — | Met |
| Case 36 | Small cell carcinoma, Lung | 62 | M | TBNA-EBUS guided, lymph node | Stage IV | Mediastinal lymphadenopathy & spinal lesions at the time of presentation | Met |
| Case 37 | Small cell carcinoma, Lung | 57 | F | Pericardial fluid | Stage IV | Brain metastasis at the time of diagnosis | Met |
| Case 38 | Small cell carcinoma, Lung | 63 | M | TBNA-EBUS guided, lymph node | Stage IV | Brain metastasis at the time of diagnosis | Met |
| Case 39 | Small cell carcinoma, Lung | 71 | M | TBNA-EBUS guided, lymph node | — | Confined to chest cavity at the time of diagnosis | Met |
| Case 40 | Small cell carcinoma, Lung | 76 | F | TBNA-EBUS guided, right hilar mass | T2N1 | Confined to chest cavity at the time of diagnosis | Primary |
| Case 41 | Poorly differentiated lung carcinoma | 50 | F | TBNA-EBUS guided, lymph node | Stage IV | Adrenal, hilar lymph nodes, and brain metastasis at the time of diagnosis | Met |
| Case 42 | Poorly differentiated lung carcinoma | 54 | F | TBNA-EBUS guided, lymph node | Stage IV | Mediastinal lymph nodes, brain, and spinal cord metastasis at the time of diagnosis | Met |
| Case 43 | Poorly differentiated lung cancer | 64 | M | TBNA-EBUS guided, RUL lung mass | Stage IIIC | — | Primary |
| Case 44 | Adenocarcinoma unknown origin | 56 | F | Pleural fluid | Stage IV | Brain and spinal cord metastasis at the time of diagnosis. | Met |
| Case 45 | Adenocarcinoma unknown origin | 59 | F | Pleural fluid | Stage IV | Positive lymph nodes and pancreatic lesion at diagnosis; presumed primary was lung mass | Met |
| Case 46 | Adenocarcinoma unknown origin | 60 | M | Ascitic fluid | — | Malignant ascites on presentation | Met |
| Case 47 | Adenocarcinoma unknown origin | 49 | F | Ascitic fluid | Stage IV | Omentum lesions and malignant ascites on presentation | Met |
| Case 48 | Adenocarcinoma unknown origin | 49 | M | Pleural fluid | Stage IV | Liver and bilateral lung lesions on presentation | Met |
| Case 49 | Mucinous adenocarcinoma unknown primary | 70 | F | Pleural fluid | Stage IV | Spinal metastasis at presentation | Met |
| Case 50 | High-grade serous ovarian adenocarcinoma | 46 | F | Ascitic fluid | pT3cpN1b | — | Met |
| Case 51 | High-grade serous ovarian adenocarcinoma | 86 | F | Pleural fluid | Stage IVA | Pelvic lymph nodes positive with abdominal and peritoneal implants on presentation | Met |
| Case 52 | High-grade serous ovarian adenocarcinoma | 65 | F | EUS FNA, gastric mass | Stage IV | Vaginal wall invasion and stomach and spleen lesions at the time of diagnosis | Met |
| Case 53 | High-grade serous ovarian adenocarcinoma | 56 | F | Pleural fluid | Stage IV | Lung and Liver metastasis at the time of diagnosis | Met |
| Case 54 | High-grade serous ovarian adenocarcinoma | 77 | F | Ascitic fluid | Stage III | Omentum and abdominal wall lesions at the time of diagnosis | Met |
| Case 55 | High-grade serous ovarian adenocarcinoma | 59 | F | Pleural fluid | Stage IV | Omentum lesions at the time of diagnosis | Met |
| Case 56 | High-grade serous ovarian adenocarcinoma | 68 | F | Pleural fluid | Stage IIIC | Carcinomatosis at the time of diagnosis | Met |
| Case 57 | High-grade serous ovarian adenocarcinoma | 60 | F | Ascitic fluid | Stage IIIC | Carcinomatosis at the time of diagnosis | Met |
| Case 58 | High-grade serous ovarian adenocarcinoma | 49 | F | Pleural fluid | Stage IV | Abdominal wall implants at the time of diagnosis | Met |
| Case 59 | Low-grade serous ovarian carcinoma | 41 | F | Pelvic washing | Stage IIIB, pT3bpN1a | — | Met |
| Case 60 | Endometrial adenocarcinoma | 60 | F | Ascitic fluid | Stage IV, pT3apN0pM1 | Serous endometrial carcinoma | Met |
| Case 61 | Endometrial adenocarcinoma | 49 | F | Ascitic fluid | Stage IV | Presented with carcinomatosis | Met |
| Case 62 | Endometrial adenocarcinoma | 78 | F | Ascitic fluid | Stage IV | Clear cell endometrial carcinoma, presented with carcinomatosis | Met |
| Case 63 | Endometrial adenocarcinoma | 59 | F | Ascitic fluid | Stage IV | Serous endometrial carcinoma | Met |
| Case 64 | Endometrial adenocarcinoma | 58 | F | FNA, left lower back soft tissue mass | Stage IV | Serous endometrial carcinoma; back mass and small intestine mass at the time of diagnosis | Met |
| Case 65 | Endometrial adenocarcinoma | 53 | F | Pelvic washing | Stage IV | Endometrioid adenocarcinoma with lung and diaphragm metastasis at the time of diagnosis | Met |
| Case 66 | Endometrial adenocarcinoma | 62 | F | Ascitic fluid | Stage IVB, pT3pM1 | Serous endometrial carcinoma with lung metastasis at presentation | Met |
| Case 67 | Endometrial adenocarcinoma | 64 | F | Pleural fluid | Stage IV | Pleural effusion and lung metastasis at the time of presentation | Met |
| Case 68 | Cervical SqCC | 62 | F | FNA, left supraclavicular mass | Unknown | Original tumor treated at outside hospital. Presented 10 years after original diagnosis with positive lymph node | Met |
| Case 69 | Cervical SqCC | 59 | F | Ascitic fluid | Stage IV | Hx of recurrent low saag ascites, Hx of Cervical cancer | Met |
| Case 70 | Cervical adenocarcinoma | 37 | F | Ascitic fluid | Stage IIB | History of treatment at outside hospital. Presented with omentum lesions and supraclavicular lymph node positive. | Met |
| Case 71 | Breast adenocarcinoma | 55 | F | Pleural fluid | Stage IV | Bone, lung, brain, and gastric metastasis at the time of diagnosis | Met |
| Case 72 | Breast adenocarcinoma | 65 | F | Pleural fluid | Unknown | Diagnosis at another institution more than 15 years before presentation. Presented to our institution with lung and liver metastasis | Met |
| Case 73 | Bilateral breast adenocarcinoma | 57 | F | Pleural fluid | pT1c | Presented more than 10 years after initial diagnosis with spine and brain metastasis and pleural effusion | Met |
| Case 74 | Breast adenocarcinoma | 48 | F | Pleural fluid | Stage IIB | Cytologic sample 4 years after original diagnosis of lung metastasis | Met |
| Case 75 | Breast adenocarcinoma | 56 | F | FNA, left breast mass | at Least Stage III | Positive lymph nodes at the time of presentation | Primary |
| Case 76 | Breast adenocarcinoma | 62 | F | Ultrasound-guided FNA, liver | Unknown | Presented to our institution 12 years after initial diagnosis with spine metastasis | Met |
| Case 77 | Breast adenocarcinoma | 73 | F | TBNA-EBUS guided, RLL mass | Unknown | Hx of breast cancer with unknown time of diagnosis, presented with lung nodule | Met |
| Case 78 | Pancreatic adenocarcinoma | 91 | M | EUS-guided FNA, pancreatic body | Stage IV | Liver and lung metastasis at the time of diagnosis | Primary |
| Case 79 | Pancreatic adenocarcinoma | 68 | F | EUS-guided FNA, pancreas | Stage IA | — | Primary |
| Case 80 | Pancreatic adenocarcinoma | 63 | M | EUS-guided FNA, liver | Stage IV | Liver and lymph node metastasis at the time of diagnosis | Met |
| Case 81 | Pancreatic adenocarcinoma | 79 | F | EUS-guided FNA, liver | Stage IV | Liver lesions at the time of diagnosis | Met |
| Case 82 | Pancreatic carcinoma with squamoid features | 64 | F | EUS-guided FNA, pancreas | Stage IV | Liver lesions at the time of diagnosis | Primary |
| Case 83 | Pancreatic adenocarcinoma | 75 | F | EUS-guided FNA, pancreatic head | Stage IV | Liver and lung metastasis at the time of diagnosis | Primary |
| Case 84 | Pancreatic adenocarcinoma | 71 | F | EUS-guided FNA, pancreas | Unknown | Only seen for diagnostic procedure | Primary |
| Case 85 | Pancreatic adenocarcinoma | 59 | M | EUS-guided FNA, pancreatic head | T3 | — | Primary |
| Case 86 | Pancreatic adenocarcinoma | 58 | M | EUS-guided FNA, pancreas | Unknown | — | Primary |
| Case 87 | Gastrointestinal stromal tumor (GIST) | 45 | F | EUS-guided FNA, stomach | pT2pN0 | — | Primary |
| Case 88 | Gastrointestinal stromal tumor (GIST) | 75 | M | EUS-guided FNA, splenic hilum | Unknown | — | Primary |
| Case 89 | Gastrointestinal stromal tumor (GIST) | 61 | F | EUS-guided FNA, mediastinal mass | Stage IV | Liver metastasis at the time of diagnosis | Primary |
| Case 90 | Gastrointestinal stromal tumor (GIST) | 59 | M | EUS-guided FNA, gastric mass | Unknown | — | Primary |
| Case 91 | Gastrointestinal stromal tumor (GIST) | 67 | M | EUS-guided FNA, stomach | pT2 | — | Primary |
| Case 92 | Gastrointestinal stromal tumor (GIST) | 48 | M | EUS-guided FNA, abdominal mass | Unknown | Patient diagnosed ten years earlier than the presentation. FNA performed on recurrence | Primary |
| Case 93 | Extrahepatic cholangiocarcinoma | 52 | M | EUS-guided FNA, pancreas | Stage IIIA | — | Primary |
| Case 94 | Cholangiocarcinoma | 82 | F | Ascites fluid | Stage IV | Malignant ascites at the time of diagnosis | Met |
| Case 95 | Gastric adenocarcinoma | 67 | F | Ascites fluid | Stage IV | Lymph nodes positive and malignant ascites at the time of diagnosis | Met |
| Case 96 | Gastric adenocarcinoma | 73 | M | Pleural fluid | Stage IIIA, pT2pN3a | — | Met |
| Case 97 | Low-grade appendiceal mucinous tumor (LAMN) | 52 | M | Peritoneal fluid | pT4b pN2 | Liver lesions and peritoneal implants at the time of presentation | Met |
| Case 98 | Head & Neck SqCC | 58 | M | Pleural fluid | Unknown | Original tumor treated at outside institution. Presented with pleural effusion and lung nodule. | Met |
| Case 99 | EBV-associated nasopharyngeal carcinoma | 56 | M | FNA, left neck mass | pT2pN2b | Lymph Node positivity at time of diagnosis | Met |
| Case 100 | Laryngeal SqCC | 54 | M | FNA, RUL mass | pT4apN0 | Hx of laryngectomy for SqCC; Biopsy of lung mass found to be metastasis | Met |
| Case 101 | Left frontal sinus SqCC | 49 | M | TBNA-EBUS guided, lymph node | pT4apN0pM0 | Hx of sinus carcinoma. FNA of lung metastasis | Met |
| Case 102 | Tongue/tonsil/FOM SqCC | 63 | M | FNA, right neck mass | Stage IVB | — | Met |
| Case 103 | SqCC of face (skin) | 87 | M | FNA, submandibular region, left | pT2pN0 | Hx of Mycosis Fungoides | Met |
| Case 104 | SqCC of larynx | 64 | F | FNA, right neck mass | Stage IVA; pT2pN2b | — | Met |
| Case 105 | Conjunctival SqCC | 78 | F | FNA, right preauricular region | pT4pN1 | Neck lymph node FNA | Met |
| Case 106 | Metastatic papillary thyroid carcinoma | 66 | F | TBNA-EBUS guided, posterior tracheal mass | Stage III, pT4 pN1a | Current presentation after treatment with radioactive iodine and resection of thyroid | Met |
| Case 107 | Papillary thyroid Carcinoma | 27 | F | US-guided FNA, left cervical lymph node | — | — | Met |
| Case 108 | High-grade salivary carcinoma | 74 | M | TBNA-EBUS guided, lymph node | At least Stage III | Hx of salivary duct carcinoma; mediastinal lymph node positive | Met |
| Case 109 | Melanoma | 38 | M | Ascites fluid | Unknown | Presented 5 year after original diagnosis with malignant ascites and spine lesions | Met |
| Case 110 | Melanoma | 43 | M | Peritoneal fluid | Unknown | Presented 12 years after original diagnosis with metastatic bone lesions | Met |
| Case 111 | Metastatic renal cell carcinoma | 57 | M | Right rib fluid | Unknown | Prior outside treatment of original tumor. Presented with bone metastasis | Met |
| Case 112 | Oncocytic renal cell carcinoma | 65 | M | CT-guided FNA, right kidney | Stage I, kidney confined lesion. | Hx of right lung cancer, radioablated renal mass; no resection performed | Primary |
| Case 113 | Urothelial adenocarcinoma | 64 | M | CT-guided FNA, Left acetabulum | pT3apN0 (Bladder tumor); Stage IV (DLBCL) | Hx of Diffuse large B cell lymphoma with prior treatment and BCG treatment of urothelial carcinoma before resection. This sample was at the time of recurrence in bone | Met |
| Case 114 | Malignant mesothelioma | 65 | M | Pleural fluid | — | — | Met |
| Case 115 | Pancreatic neuroendocrine tumor | 71 | M | EUS-guided FNA, pancreas | — | — | Primary |
| Case 116 | Neuroendocrine tumor, duodenum | 66 | F | EUS-guided FNA, duodenal bulb | Stage IV; pT3pN1pM1 | Liver metastasis at the time of diagnosis | Primary |
| Case 117 | Neuroendocrine tumor | 77 | F | EUS-guided FNA, ampulla | Stage IV; pM1 | Spine and liver metastatic lesions at the time of diagnosis | Met |
A total of 20 negative controls were identified in FNA lymph node samples; 10 negative lymph nodes from patients with no history of malignancy, and 10 negative lymph nodes from patients with a history of malignancy were included.
2.2 DNA and RNA extraction
FFPE cytology cell blocks were sectioned at 7 μm. A total of 15 unstained and unbaked serial sections were used for both DNA and RNA extraction. A 5 μm section was stained with hematoxylin and eosin. Tumor area was marked, and the percentage of tumor cells was estimated by a pathologist. Tumor cells were manually macrodissected to enrich for tumor fraction to achieve ≥20% tumor cells in the DNA/RNA sample. The Promega semi-automated FFPE DNA and RNA extraction kits were used with the Promega Maxwell RSC system (Promega Corporation). Quantitation of DNA and RNA by Qubit (Thermo Fisher Scientific) was performed prior to library preparation.
2.3 Targeted panel sequencing
Library preparation and sequencing using the Oncomine Comprehensive Assay version 3 was performed on all cases, using 20 ng of DNA and 20 ng of RNA for each sample, as has been previously described.6 The assay targets 161 unique genes including 84 genes for hotspot mutations, 43 genes for focal copy number gains, 48 full coding sequences for deletion mutations, and 51 fusion drivers. Data analysis was performed using the Torrent Suite software version v5.10. Ion Reporter version v5.10 was used for variant calling. A minimum average depth of coverage of 600X was considered adequate for each sample. Fusion analysis was performed with Ion Reporter version v5.10 fusion analysis workflow. Variants were classified as benign, likely benign, variant of undetermined significance, likely pathogenic or pathogenic based on the clinical criteria set by the College of American Pathologists and the Association of Molecular Pathology.7
2.4 Statistical analysis of outcome data
For cytologic and surgical comparison, results from a total of 17 paired samples were included. For each subject, the locus, genes, AA change, and values for variants (surgical and cytology) were recorded. The goal of this analysis was to determine the correlation between variants detected in surgical and cytology samples. Pearson's and Spearman's correlations were measured between variants. Parametric paired t-test was performed to compare the means of variants. Nonparametric paired test (Wilcoxon Signed Rank Test) was used to compare the medians of variants. The significance levels were set at 0.05 for all tests. The SAS 9.4 Version (SAS Institute, Inc.) was used for data management and analyses.
A clinical chart review was performed for all cases that passed quality metrics. A total of 117 subjects were included in this analysis. The charts were reviewed for the following information: date of diagnosis, final pathology diagnosis, treatment regimen, date of recurrence, date of metastasis diagnosis, date of death, and date of last known contact. Time of overall survival (OS) was calculated as the time from study enrollment to death or last contact. Time of progression-free survival (PFS) was calculated as the time from study enrollment to disease progression date, death date, or last contact whichever comes first. The survivor functions for PFS or OS were estimated by Kaplan-Meier survival analysis. Cox proportional hazards model was employed to estimate the adjusted effect of variants, diagnosis, and metastasis status on PFS or OS after adjustment for all other factors.
3 RESULTS
3.1 Case cohort
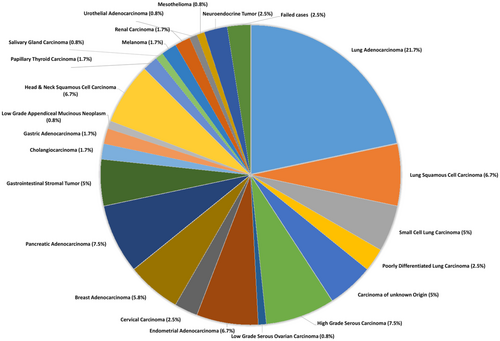
A total of 120 cases met the criteria for inclusion in the study. Two samples did not yield enough DNA for library preparation, and one case had too much formalin-induced artifacts for reliable analysis, leaving 117 samples in our study. There were 31 primaries, 6 recurrences, and 80 metastatic cases included in the study (Table 1), and the percentage of tumor cells ranged from 20–100%. The largest portion of the cohort consisted of lung carcinomas (35.8%) including 26 adenocarcinomas, 8 squamous cell carcinomas (SCCs), 6 small cell carcinomas, and 3 poorly differentiated carcinomas, which adequately reflects the frequency of thoracic specimens sent to our university's cytology service. The remaining cases consisted of 6 carcinomas of unknown origin, 9 high-grade serous carcinoma (HGSC) of the ovary and fallopian tube, 1 low-grade serous carcinoma (LGSC), 8 endometrial carcinomas (ECs) (including endometrioid, serous, and clear cell subtypes), 3 cervical carcinoma (1 squamous and 2 adenocarcinoma), 7 breast adenocarcinoma, 9 pancreatic adenocarcinoma, 6 gastrointestinal stromal tumors (GIST), 2 cholangiocarcinoma, 2 gastric adenocarcinoma, 1 low-grade appendiceal mucinous tumor, 7 head and neck SCCs (including larynx, pharynx, ocular, and oral cavity), 2 thyroid carcinoma, 1 salivary gland neoplasms, 2 melanoma, 2 renal cell carcinoma, 1 mesothelioma, 1 urothelial carcinoma, and 3 well-differentiated neuroendocrine tumors. A breakdown of each tumor type is represented in the pie chart (Figure 2).
3.2 Surgical-Cytologic correlation
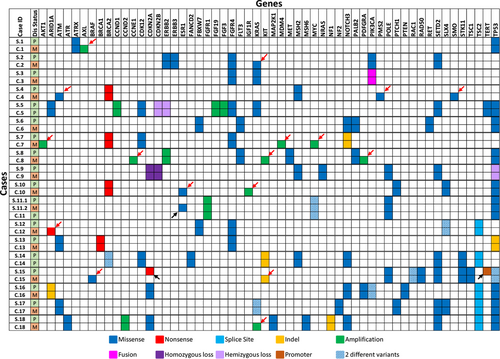
There were 18 cases within the cohort that had paired surgical and cytology samples. These samples were from different time points over the course of patient treatment (ie: surgical case at primary diagnosis and cytology case at the time of metastasis), and this difference in time was noted in our correlation results table as 1st diagnosis and 2nd diagnosis (Table 2). The Spearman correlation between 1st and 2nd diagnosis was 0.65056 with a p-value of <0.0001, and the Pearson correlation between 1st and 2nd diagnosis was 0.54763 with a p-value of <0.0001. Both methods show a significant correlation between the surgical-cytologic paired samples. A detailed comparison of variants detected in each of the surgical and cytology cases is shown in Figure 3. Ten out of 18 cases had discrepancy in variants between the surgical and cytology samples. In 8 cases, the cytology samples had extra variants compared to surgical samples. All 8 samples were from metastatic disease with additional copy number, missense, or nonsense variants that are likely associated with tumor progression and/or metastasis. In case 15, the surgical sample had a nonsense mutation (R58*) in CDKN2A (VAF 74.9%) and a promoter mutation (c.-146C > T) in TERT (VAF 50.7%) that were not present in the cytology sample. The cytology sample had an indel (W557_K558del) in KIT (VAF 11.3%) and a BRAF V600E (VAF 5.3%) mutation that were not detected in the surgical sample (Figure 3). It is possible that the cytology sample represents a different clone that expanded in the tumor metastasis. Finally, for case 11, there were two surgical samples and one cytology sample. The first surgical sample and cytology samples were both from the primary breast cancer, whereas the second surgical sample was from the metastasis. In this case, the second surgical sample had a pathogenic ESR1 D538G mutation known to be associated with endocrine therapy resistance in breast cancer.8 The rest of the variants were identical in both surgical and cytology samples (Figure 3).
| Case # | Variants at 1st diagnosis | Variants at 2nd diagnosis |
|---|---|---|
| 1 | TP53 p.Y220C; ATRX p.N1860S | TP53 p.Y220C; ATRX p.N1860S; AXL amplification |
| 2 | PIK3CA p.E542K; ERBB3 p.V104L; ERBB2 p.R678Q; SETD2 p. P1962L; SETD2 p.M1080I; FGFR4 p.G388R | PIK3CA p.E542K; ERBB3 p.V104L; ERBB2 p.R678Q; SETD2 p. P1962L; SETD2 p.M1080I; FGFR4 p.G388R; KRAS p.G12V |
| 3 | KRAS p.G12V; FGFR4 p. G388R; FNDC3B-PIK3CA fusion | KRAS p.G12V; FGFR4 p. G388R; FNDC3B-PIK3CA fusion |
| 4 | MET p. T1010I; BRCA2 p. L3326* | MET p. T1010I; BRCA2 p. L3326*; PMS2 p.H479Q; SMO p.V270I; ATM p.L1420F |
| 5 | KRAS p.G12V; TP53 p.L132R; ARID1A p.P120S; SETD2 p.P1962L; TERT p.A279T; FGFR4 p.G388R; CDK12 p.L1189Q; CCND1 Amplification; FGF19 amplification; FGF3 amplification; CDKN2A hemizygous loss; CDKN2B hemizygous loss | KRAS p.G12V; TP53 p.L132R; ARID1A p.P120S; SETD2 p.P1962L; TERT p.A279T; FGFR4 p.G388R; CDK12 p.L1189Q; CCND1 Amplification; FGF19 amplification; FGF3 amplification; CDKN2A hemizygous loss; CDKN2B hemizygous loss |
| 6 | FBXW7 p.R465Ll TP53 p.G244S; RET p.E867D; FLT3 p.I417L; PALB2 p.P210L; NOTCH3 p.A1020P | FBXW7 p.R465Ll TP53 p.G244S; RET p.E867D; FLT3 p.I417L; PALB2 p.P210L; NOTCH3 p.A1020P |
| 7 | BRCA2 p.R2318*; NOTCH3 p.Y1106fs; FGFR4 p.G388R; CDK12 p.I1131V | BRCA2 p.R2318*; NOTCH3 p.Y1106fs; FGFR4 p.G388R; CDK12 p.I1131V; MDM4 amplification, MYC amplification, AKT1 amplification |
| 8 | TP53 p.G244V; MET p.I316M; FLT3 p.A988P; PALB2 p.P210L; NOTCH3 p.A1020P; ERBB2 amplification | TP53 p.G244V; MET p.I316M; FLT3 p.A988P; PALB2 p.P210L; NOTCH3 p.A1020P; ERBB2 amplification; PDGFRA amplification; KIT amplification; CNNE1 amplification |
| 9 | NRAS p.Q61R; MSH2 p.L449N; SETD2 p.P1962L; POLE p.R1556W; CDKN2A homozygous loss; CDKN2B homozygous loss; TP53 hemizygous loss | NRAS p.Q61R; MSH2 p.L449N; SETD2 p.P1962L; POLE p.R1556W; CDKN2A homozygous loss; CDKN2B homozygous loss; TP53 hemizygous loss |
| 10 | BRCA2 p.L3326*; TP53 p.R282W; PTCH1 p.T728M; SLX4 p.R1761C | BRCA2 p.L3326*; TP53 p.R282W; PTCH1 p.T728M; SLX4 p.R1761C; ESR1 p.D538G; IGF1R amplification |
| 11 |
1st Surgery: FGFR1 amplification, MYC amplification, MYC p.V185I, POLE P697S, TP53 splice site; 2nd Surgery: ESR1 p.D538G, FGFR1 amplification, MYC amplification, MYC p.V185I, POLE P697S, TP53 splice site |
FGFR1 amplification, MYC amplification, MYC p.V185I, POLE P697S, TP53 splice site |
| 12 | FBXW7 p.R479P; FGFR4 p.G388R; SLX4 p.T919I; SLX4 p.W546C; TSC2 splice site | ARID1A p.E1718*; FBXW7 p.R479P; FGFR4 p.G388R; SLX4 p.T919I; SLX4 p.W546C; TSC2 splice site |
| 13 | TP53 p.R248fs; BRCA1 p.Q1806*; FGFR4 p.G388R; ATM p.B122T | TP53 p.R248fs; BRCA1 p.Q1806*; FGFR4 p.G388R; ATM p.B122T |
| 14 | MSH2 p.I770V; FAND2 p.Q65H; SETD2 p.P1962L; BRCA2 p.H1561N; BRCA2 p.V2138F; CDK12 p.T1195M; STK11 p.F354L; NOTCH3 p.A1020P; TSC2 splice site; KIT deletion | MSH2 p.I770V; FAND2 p.Q65H; SETD2 p.P1962L; BRCA2 p.H1561N; BRCA2 p.V2138F; CDK12 p.T1195M; STK11 p.F354L; NOTCH3 p.A1020P; TSC2 splice site; KIT deletion |
| 15 | RAC1 p.P29F; RAC1 p.P29L; CDKN2A p.R58*; TP53 p.E286K; TP53 p.A159V; SETD2 p.P1962L; RAD50 p.D767N; TSC1 p.K587R; POLE p.G6R; STK11 p.D350N; TERT promoter | RAC1 p.P29F; RAC1 p.P29L; TP53 p.E286K; TP53 p.A159V; SETD2 p.P1962L; RAD50 p.D767N; TSC1 p.K587R; POLE p.G6R; STK11 p.D350N; BRAF p. V600E; KIT p.W557_K558del |
| 16 | ARID1A p.V1817fs; PIK3CA p.E81K; PIK3CA p.R88Q; PTEN p.Y68H; TP53 p.R273H; CDKN2A p.H123Q; PALB2 p.T386A; NOTCH3 p.A1020P; TSC2 splice site | ARID1A p.V1817fs; PIK3CA p.E81K; PIK3CA p.R88Q; PTEN p.Y68H; TP53 p.R273H; CDKN2A p.H123Q; PALB2 p.T386A; NOTCH3 p.A1020P; TSC2 splice site |
| 17 | KRAS p.G12F; KRAS p.G12C; TP53 p.R273L; SETD2 p.P1962L; PTCH1 p.T728M; ATM p.M1040V; SLX4 p.P975L; NF2 p.E463L; TSC2 In/Del | KRAS p.G12F; KRAS p.G12C; TP53 p.R273L; SETD2 p.P1962L; PTCH1 p.T728M; ATM p.M1040V; SLX4 p.P975L; NF2 p.E463L; TSC2 In/Del |
| 18 | CDKN2A p.D108H; MAP2K1 p.P124L; TP53 p.L194R; NF1 p.Y80fs; MSH6 p.S63P; SETD2 p.P1962L; ATR p.S1607N; NOTCH3 p.A1020P; TSC2 splice site; CCND2 amplification | CDKN2A p.D108H; MAP2K1 p.P124L; TP53 p.L194R; NF1 p.Y80fs; MSH6 p.S63P; SETD2 p.P1962L; ATR p.S1607N; NOTCH3 p.A1020P; TSC2 splice site; CCND2 amplification; KRAS amplification |
- Note: Bolded text identifies variants not identified in both specimens.
3.3 Cytology cases
Within the 117 cases sequenced, the depth of coverage ranged from 648X to 2694X (Figure 4A). A total of 711 variants were detected, including 505 single nucleotide variants (SNVs), 2 multinucleotide variants (MNVs), 73 insertion/deletions (indels), 126 copy number variants (CNVs), and 5 fusions (Figure 4B, Figure 5, Table S1). There was a total of 347 pathogenic variants and 345 variants of undetermined significance (VUS) detected in all cases (Figure 4C). The 20 negative control samples were sequenced with adequate depth of coverage and no pathogenic variants were detected.
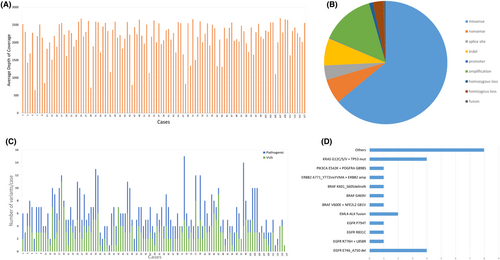

In the entire cohort of 117 cases, TP53 was the most frequently mutated (58%) gene (Figure 5). Among the 26 lung adenocarcinomas, 18 cases had clinically actionable variants for which FDA-approved drugs are available. Seven cases had EGFR mutations (4 cases with exon 19 deletion, 1 case with R776H and L858R, 1 case with R831C, and 1 case with P794T). Four cases with KRAS G12C/S/V and TP53 variants, 2 cases with EML4-ALK fusion, 3 cases with BRAF variants (1 V600E, 1 G469V, and 1 exon 15 indel), 1 case with PIK3CA E542K, and 1 case with ERBB2 exon 20 indel. In the remaining 8 cases variants in NF1, STK11, ERBB4, SETD2, BAP1, and CDKN2A genes were detected (Figure 4D, Table S1). One case with KRAS G12S also had a WHSC1L1-FGFR1 fusion. The squamous cell lung carcinomas showed two cases with ARID1A loss of function variants (E1718* and G1848fs), one FGFR3 S249C variant, one MAP2K1 P124L, and one case with PIK3CA amplification and E545A variant. One case of poorly differentiated carcinoma of the lung had an ARID1A G836fs variant. The small cell carcinoma cases and remaining lung carcinoma cases did not have clinically relevant pathogenic variants.9
Of the 6 carcinomas of unknown origin, 2 cases showed a KRAS G12D/V variant and one case showed an amplification of the same gene. One case had an ERBB2 amplification with a SMARCA4 loss of function G1232C variant. One case had many amplifications present.
Our cohort of gynecologic tumors included 9 high-grade serous ovarian/fallopian tube cancers (HGSC), one LGSC, 8 ECs, one SCC of the cervix, and two adenocarcinomas of the cervix. Of the HGSC, two cases had a BRCA1 pathogenic variant (W1733* and Q1806*), and one case had a BRCA2 pathogenic variant (L1227fs). Additionally, one case showed a FGFR2 amplification and another case showed a FGFR1 pathogenic variant (D166del). The LGSC showed a PIK3CA-FNDC3B fusion and a KRAS G12V variant. Of the 8 ECs, two had the most common hotspot PIK3CA variants (Q546K and E542A), one case with two different hotspot PIK3CA variants (E81K and R88Q) and an additional case with a PIK3CA amplification. One case of EC had a BRCA1 frameshift variant (E1210fs), and one case showed ERBB2 amplification. FGFR3 amplification and gain-of-function variant in one case appeared to be the driving alterations. Within the 7 breast cancer cases, two cases showed a BRCA2 nonsense variant (p.R2318* and p.K3326*), one case showed ERBB3 G234R, one case showed KRAS G12V, and one case showed ARID1A P453fs. No PIK3CA, BRCA1, or ERBB2 alterations were identified in the breast carcinoma cases.10, 11 Amplifications of AKT1, MYCN, MYC, FGFR1, FGFR4, FGF3, FGF19, IGF1R, CCND1, CCNE1, TERT, and NTRK3 were detected in 4/7 cases (Figure 5).
All nine pancreatic adenocarcinomas harbored exon 2 pathogenic gain-of-function variants in KRAS and 6/9 cases had a deleterious loss-of-function variant in TP53. In addition to these variants, one case showed a frameshift mutation in BRCA1 (Q867fs), and another case showed a less common G1049R gain-of-function variant in PIK3CA. All six cases of GIST harbored a pathogenic variant in KIT—three indels including a splice site variant and 3 gain of function missense variants. Of the two cholangiocarcinoma cases, one case had a SMAD4 R361H pathogenic variant and the other case had a pathogenic KRAS, TP53, and additional CTNNB1 S45F variant. The two gastric carcinoma cases showed an ERBB2 R678Q pathogenic variant with a PIK3CA E542K and KRAS G12V variants.
Within the SCCs of the head and neck region, there was variability in locations that was reflected in the variants detected. One case from the face showed two loss of function variants in BRCA1 (E17758* and E1491*). A frontal sinus-based neoplasm had a loss of function BRCA2 K3326* variant. A nasopharyngeal-based lesion harbored a pathogenic HRAS Q61R variant and a FGFR1 amplification. A laryngeal SCC was found to have a PIK3CA H1047R pathogenic variant. The additional larynx-based tumor and the floor of mouth tumor included in our cohort showed significant number of copy number variations (CNVs).12
In the smaller diagnosis cohort, one melanoma case had a NRAS Q61R variant and the other case of melanoma had a BRAF V600E variant. Both papillary thyroid carcinoma cases harbored BRAF V600E variants. One case of renal cell carcinoma harbored a KRAS G12D variant and showed an oncocytic papillary phenotype. The salivary gland malignancy showed PIK3CA H1047R and HRAS Q61R pathogenic variants.
Of note, there were no significant pathogenic variants detected in the urothelial carcinoma, the mesothelioma, the low-grade appendiceal mucinous tumor, and neuroendocrine tumors. A summary of variants along with diagnosis and clinical relevance is shown in Figure 5, and for a full list of variants detected in our cohort, please refer to Table S1.
4 DISCUSSION
In recent years, cytology samples are a growing proportion of samples that are sent for routine molecular diagnostics. Our study demonstrates that NGS-based targeted sequencing can be performed in a highly accurate and reproducible manner with small quantity cytology samples. Within our university cytology service, we found that approximately 27% (120/451) of our malignant cytologic samples had cell block material adequate for molecular testing via a NGS comprehensive assay. Of the 117 cases included in this study, 18 cases had a surgical pathology sample available and showed good concordance of results between cytologic and surgical pathology specimens validating the utility of cytologic specimens in the molecular diagnostics laboratory.
One of the challenges of using comprehensive NGS-based assays for routine diagnostic testing is the quality and quantity of tissue required for detecting clinically relevant mutations. Our study demonstrates that we had a very small failure rate (2.5%; 3/120) due to either sample size or formalin-induced artifacts. Our assay detected variants (SNVs) with 5% or higher allele frequency, CNVs (amplification, single copy loss and homozygous loss) and gene fusions efficiently. For many tumors, the percent tumor cells present in the section amounted to 20% but we were able to enrich the tumor fraction by macrodissecting to yield sufficient DNA and RNA for running the assay successfully.
Genomic heterogeneity plays a significant role in eventual drug resistance and treatment failure resulting from the generation of subpopulations within a tumor. The Cancer Genome Atlas (TCGA) studies performed by inter- and intra-tumor comparisons have shown tumor heterogeneity in many types of tumors including lung, breast, prostate, glioblastoma and colon cancers.10, 13-16 Furthermore, TCGA pan-cancer analysis of genomic landscapes of 12 tumor types from more than 3000 tumors identified 127 significantly mutated genes with both established and emerging links to cancer, indicating that the number of driver mutations required for oncogenesis is relatively small.17 Although a common set of driver mutations exists in a given cancer type, the combination of mutations within a patient tumor and their distribution within the founding clone and subclones will be critical for optimizing their treatment. We sequenced the samples at a higher depth of coverage to account for the sub-populations of cells that might be contributing toward the makeup of the tumor.
One of the limitations with cytology samples is the small quantity of cellular material, which raises concerns about capturing tumor heterogeneity. In FNA biopsy samples, multiple planes of the tumor and the resulting specimen comprises multiple passes through the tumor. In body fluid specimens, concern for heterogeneity should be low because malignant cells in cavity fluids typically represent the most aggressive and metastatic subclone of the tumor and would be the ideal subpopulation to study. Methodologically, increasing the percent of tumor cells in a sample by selective microdissection proportionally increases the yield of tumor cells coming from the major subpopulations, thereby increasing the yield of both driver and passenger mutations.18
In our cohort, we identified 32 cases (27.4%) that would qualify for FDA-approved targeted therapy according to the current guidelines. An additional 23 cases (19.7%) would qualify for a clinical trial based on pathogenic variants detected by NGS (Table 3). With approximately 45% of cases in our cohort qualifying for some form of targeted therapy, the critical importance of providing high complexity NGS testing of cytologic samples is undeniable. Currently available treatments include imatinib for KIT alterations in GIST, gefitinib, osimertinib, sotorasib, crizotinib, for alterations in EGFR, KRAS, and ALK fusion in NSCLC, PARP inhibitors, and alpelisib for BRCA1/2 and PIK3CA variants in gynecologic malignancies, and dabrafenib and trametinib for BRAF V600E in melanoma and metastatic papillary thyroid carcinoma.19-24Our cohort does not include enough samples for each diagnosis to reach statistical significance with regard to clinical outcome. But comparing cases with and without actionable variants in our overall cohort, there was a significant correlation between cases with targetable variants with both PFS and OS (Figure S1).
| Case ID | Diagnosis | Clinical/Pathologic Stage at Diagnosis | Primary/Metastasis | Gene | Exon | Variant type | AA Change | FDA-Approved Therapy Options | Clinical Trial Options |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Lung Adeno | Stage IVB; pT4pM1c | Met | EGFR | 19 | INDEL | p.E746_A750del | Yes | |
| 2 | Lung Adeno | pT2apN0 | Recurrence | KRAS | 2 | SNV | p.G12C | Yes | |
| 4 | Lung Adeno | Stage IVB | Met | EGFR | 19 | INDEL | p.E746_A750del | Yes | |
| 5 | Lung Adeno | Stage IV | Met | ERBB2 | 20 | INDEL | p.A771_Y772insYVMA | Yes | |
| 6 | Lung Adeno | Stage IIIA | Met | BRAF | 15 | SNV | p.V600E | Yes | |
| 11 | Lung Adeno | Stage IVB | Met | EML4-ALK | e13|e20 | FUSION | Read count: 27218 | Yes | |
| 12 | Lung Adeno | Stage IV | Primary | EGFR | 21 | SNV | p.L858R | Yes | |
| 14 | Lung Adeno | Stage IVA | Primary | PIK3CA | 10 | SNV | p.E542K | Yes | |
| 15 | Lung Adeno | Stage IV | Met | BRAF | 11 | SNV | p.G469V | Yes | |
| 16 | Lung Adeno | Unknown | Primary | EGFR | 19 | INDEL | p.E746_A750del | Yes | |
| 17 | Lung Adeno | Stage IVB; pT2b pN3 pM1c | Primary | EML4-ALK | e13|e20 | FUSION | Read count: 131144 | Yes | |
| 20 | Lung Adeno | Stage I, pT1 pN0 | Recurrence | NF1 | 37 | INDEL | p.C1682Ter | Yes | |
| 21 | Lung Adeno | Stage IVB | Met | EGFR | 19 | INDEL | p.E746_A750del | Yes | |
| 22 | Lung Adeno | At least Stage III | Met | EGFR | 21 | SNV | p.R831C | Yes | |
| 23 | Lung Adeno | Stage IIIA | Primary | NF1 | 42 | INDEL | p.L2103fs | Yes | |
| 26 | Lung Adeno | Stage IV | Met | KRAS | 2 | SNV | p.G12C | Yes | |
| 27 | Lung SqCC | Stage IIIB | Recurrence | ARID1A | 20 | SNV | p.G1848fs | Yes | |
| 30 | Lung SqCC | Stage IIIB | Met | ARID1A | 20 | SNV | p.E1718* | Yes | |
| 32 | Lung SqCC | Stage IIIA | Primary | FGFR3 | 7 | SNV | p.S249C | Yes | |
| 33 | Lung SqCC | At least Stage IIIA | Primary | PIK3CA | 10 | SNV | p.E545K | Yes | |
| 41 | Poorly differentiated lung ca | Stage IV | Met | ARID1A | 8 | SNV | p.G836fs | Yes | |
| 51 | High-grade serous ovarian ca | Stage IVA | Met | BRCA2 | 11 | SNV | p.L1227fs | Yes | |
| 52 | High-grade serous ovarian ca | Stage IV | Met | BRCA1 | 18 | SNV | p.W1733* | Yes | |
| 54 | High-grade serous ovarian ca | Stage IV | Met | BRCA1 | 22 | SNV | p.Q1806Ter | Yes | |
| 57 | High grade serous ovarian ca | Stage IIIC | Met | ARID1A | 1 | SNV | p.A246fs | Yes | |
| 61 | Endometrial adenocarcinoma | Stage IV | Met | PIK3CA | 10 | SNV | p.E542A | Yes | |
| 62 | Endometrial adenocarcinoma | Stage IV | Met | ERBB2 | Amp | CNV | Copy number: 16 | Yes | |
| 63 | Endometrial adenocarcinoma | Stage IV | Met | BRCA1 | 10 | SNV | p.E1210fs | Yes | |
| 64 | Endometrial adenocarcinoma | Stage IV | Met | ARID1A | 20 | SNV | p.V1817fs | Yes | |
| 65 | Endometrial adenocarcinoma | Stage IV | Met | PIK3CA | 10 | SNV | p.Q546K | Yes | |
| 66 | Endometrial adenocarcinoma | Stage IVB, pT3pM1 | Met | ERBB2 | Amp | CNV | Copy number: 8.4 | Yes | |
| 68 | Cervical SqCC | Unknown | Met | PIK3CA | 10 | SNV | p.E545K | Yes | |
| 69 | Cervical SqCC | Stage IV | Met | PIK3CA | 10 | SNV | p.E542V | Yes | |
| 71 | Breast adenocarcinoma | Stage IV | Met | ERBB3 | 7 | SNV | p.G284R | Yes | |
| 72 | Breast adenocarcinoma | Unknown | Met | ARID1A | 3 | SNV | p.P453fs | Yes | |
| 73 | Bilateral breast adenocarcinoma | pT1c | Met | BRCA2 | 13 | SNV | p.R2318* | Yes | |
| 74 | Breast adenocarcinoma | Stage IIB | Met | BRCA2 | 27 | SNV | p.K3326* | Yes | |
| 87 | Gastro intestinal stromal tumor | pT2pN0 | Primary | KIT | 11 | INDEL | p.W557_K558del | Yes | |
| 88 | Gastro intestinal stromal tumor | Unknown | Primary | KIT | 11 | SNV | p.V559G | Yes | |
| 89 | Gastro intestinal stromal tumor | Stage IV | Primary | KIT | 11 | INDEL | p.W557_K558del | Yes | |
| 90 | Gastro intestinal stromal tumor | Unknown | Primary | KIT | 11 | SNV | p.W557G | Yes | |
| 91 | Gastro intestinal stromal tumor | pT2 | Primary | KIT | 11 | SNV | p.V559G | Yes | |
| 92 | Gastro intestinal stromal tumor | Unknown | Recurrence | KIT | 11 | INDEL | Splice site | Yes | |
| 94 | Cholangiocarcinoma | Stage IV | Met | RET | 13 | SNV | p.Y791F | Yes | |
| 100 | Laryngeal SCC | pT4apN0 | Met | PIK3CA | 21 | SNV | p.H1047R | Yes | |
| 101 | Left frontal sinus SCC | pT4apN0pM0 | Met | MET | 14 | SNV | p.T1010I | Yes | |
| 102 | Tongue/tonsil/FOM SCC | Stage IVB | Met | ARID1A | 9 | SNV | p.Q944* | Yes | |
| 103 | SCC of face (skin) | pT2pN0 | Met | BRCA1 | 20 | SNV | p.E1775* | Yes | |
| 104 | SCC of larynx | Stage IVA; pT2pN2b | Met | MSH2 | 15 | SNV | p.C873* | Yes | |
| 105 | Conjunctival SCC | pT4pN1 | Met | KIT | 11 | INDEL | p.W557_K558del | Yes | |
| 106 | Metastatic papillary thyroid ca | Stage III, pT4 pN1a | Recurrence | BRAF | 15 | SNV | p.V600E | Yes | |
| 107 | Papillary thyroid ca | Unknown | Met | BRAF | 15 | SNV | p.V600E | Yes | |
| 108 | High-grade salivary ca | At least Stage III | Met | PIK3CA | 21 | SNV | p.H1047R | Yes | |
| 109 | Melanoma | Unknown | Met | BRAF | 15 | SNV | p.V600E | Yes | |
| 110 | Melanoma | Unknown | Met | NRAS | 3 | SNV | p.Q61R | Yes |
It is important to note that 50% of the cases that came through the cytology service during the period of archival review were not included in this study due to lack of material to review or due to insufficient tissue for further analysis. Because of the retrospective nature of this study, some samples had cell blocks that were exhausted or had very little tissue left that was inadequate for molecular testing. Additional prospective studies would need to be done to get a more accurate success rate of cytologic cell blocks in light of tissue preservation methods used in laboratories today to improve ancillary testing results. Alternative forms of cell preservation and additional FNA passes dedicated for molecular laboratories could also increase the number of cytologic cases for successful molecular analysis. At our institution, a high number of patients diagnosed by the cytology service as “positive for malignancy” are high stage at presentation or have recurrent disease, and these patients would benefit most from molecular analysis. Although intra-tumor heterogeneity is a concern, molecular analysis of cytologic specimens can adequately identify driver mutations for which targeted therapy options are available. As shown in our comparison of surgical samples and cytologic cell blocks, the major drivers of oncogenesis in each tumor pair were identified in both samples. As most of the patients in our cohort either were not eligible for surgical resection or had not responded to first-line chemotherapy at the time of the cytologic sample collection, this is exactly the cohort that would most benefit from approved targeted therapies or clinical trial options. With this in mind, and extrapolating from our study, approximately 30% more of the patients in our population with similar clinical histories would qualify for currently approved targeted therapy regimens or would qualify for a clinical trial. We continue to optimize our service for such patients and have developed confidence in the methods and yield of targeted sequencing of solid tumor in FNA and body fluid cytology cell block specimens.
AUTHOR CONTRIBUTIONS
Erica Vormittag-Nocito: Conceptualization (supporting); methodology (supporting); writing – original draft (supporting); writing – review and editing (supporting). Ravindra Kumar: Data curation (supporting). Kunwar Narayan: Investigation (supporting); project administration (supporting). Zhengjia Chen: Software (lead). Odile David: Project administration (supporting); resources (supporting); supervision (supporting). Frederick Behm: Resources (lead); supervision (supporting). Gayatry Mohapatra: Conceptualization (equal); data curation (equal); formal analysis (equal); project administration (equal); writing – original draft (lead); writing – review and editing (lead).
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
None.
ETHICAL APPROVAL STATEMENT
This study was approved by the University of Illinois Institutional Review Board with an exemption granted for written informed consent.
CLINICAL TRIAL REGISTRATION
N/A
Open Research
DATA AVAILABILITY STATEMENT
Data will be shared once the manuscript is accepted.