Fc-gamma receptor polymorphisms, cetuximab therapy, and overall survival in the CCTG CO.20 trial of metastatic colorectal cancer
Funding information
The study was supported by the Alan B. Brown Chair in Molecular Genomics, Princess Margaret Hospital and Ontario Cancer Institute, University of Toronto, Toronto, ON.
Abstract
Background
Two germ line Fc-γ receptor (FCGR) polymorphisms, rs1801274 [FCGR2A; His(H)131Arg(R)] and rs396991 [FCGR3A; Phe(F)158Val(V)], produce altered proteins through amino acid substitutions. We previously reported that the FCGR2A H/H genotype was associated with longer overall survival (OS) in cetuximab-treated chemotherapy-refractory patients with metastatic colorectal cancer. Here, we aimed to replicate and extend this finding in the Canadian Clinical Trials Group CO.20 trial.
Methods
After germ line DNA genotyping, polymorphic relationships with survival were assessed using log-rank tests and hazard ratios (HR) from Cox proportional hazard models, adjusting for known prognostic factors. The dominant genetic inheritance model was used for the main analysis.
Results
Of 592 wild-type KRAS patients treated with cetuximab, those with the FCGR2A H/H genotype (n = 165, 28%) had improved OS (HR: 0.66, P < 0.001; median absolute benefit, 1.3 months) compared to those with R/- genotype (n = 427, 72%). Patients with H/R had intermediate results under a codominant genetic inheritance model (HR: 0.72, P = 0.003). No significant associations were found between FCGR3A genotype and OS. In an exploratory analysis, patients with the combination of FCGR2A H/H + FCGR3A F/F genotype had significantly better OS (HR: 0.33, P = 0.003; median absolute benefit, 12.5 months) than patients with the combination of double-variant R/R + V/V genotype. Progression-free survival results were similar to OS. Toxicity rates were not associated with either polymorphism.
Conclusions
The FCGR2A genotype was associated with efficacy but not with toxicity in wild-type KRAS, cetuximab-treated colorectal cancer patients. FCGR3A genotype may modulate the relationship between FCGR2A polymorphism and outcome. FCGR2A is a promising biomarker for clinical management for these patients.
1 INTRODUCTION
Cetuximab is an IgG1 monoclonal antibody (mAb) that targets theepidermal growth factor receptor (EGFR) and has been shown to improve outcomes in patients with metastatic wild-type KRAS colorectal carcinoma.1, 2 As many of these patients do not benefit from cetuximab treatment,2 there is an unmet need for additional predictive biomarkers, in addition to RAS and BRAF mutations.
One of cetuximab's mechanisms of action is antibody-dependent cellular cytotoxicity (ADCC).3, 4 ADCC is initiated when the antigen-binding fragment (Fab) binds to the tumor cell and the crystallizable fragment (Fc) binds to the crystallizable fragment gamma receptor (FCGR) on a natural killer cell, macrophage, or monocyte, creating a bridge from the tumor cell to the effector cell. Tumor cell recognition is then coupled with a lytic attack on the cancer cell mounted by effector cells.5, 6 Three classes of FCGR exist, encoded by related genes on the long arm of chromosome 1: FCGR1-CD64; FCGR2-CD32; and FCGR3-CD16.7 Two polymorphisms located within coding regions of FCGR2A and FCGR3A were previously reported to be associated with the efficacy of cetuximab in colorectal cancer.8, 9 A nonsynonymous polymorphism in the extracellular domain of FCGR2A (rs1801274) changes the amino acid from histidine (H) to arginine (R), significantly reducing the receptor's affinity to Fc.17 The rs396991 polymorphism in FCGR3A is also found in the extracellular domain, leading either to a phenylalanine (F) or valine (V) substitution; this amino acid interacts with the lower hinge region of IgG1.18, 19 Previous studies of the association of these two polymorphisms with the efficacy of cetuximab reported mixed results.8, 9 Most of these studies had various limitations, including small sample size, non-randomized patient selection, and suboptimal genotyping technique. A recent analysis of data from the Canadian Cancer Trials Group (CCTG) CO.17 randomized controlled trial found cetuximab treatment was associated with overall survival (OS) benefit in patients with metastatic wild-type KRAS colorectal cancer who had the FCGR2A H/H genotype but not those with the R/- genotype. Patients with the H/R genotype had non-statistically significant intermediate results.20 A post hoc analysis found cetuximab-treated patients with the H/H genotype had longer OS than those with R/- genotype (univariate hazard ratio (HR) 0.63 (95% confidence interval (CI) 0.3-0.9), adjusted HR: 0.57, 95% CI: 0.3-0.8). This effect was not seen in the best supportive care arm. In contrast, no association was found between the FCGR3A polymorphism and any clinical outcome. The primary objective of this study was to replicate our previous finding of the association of FCGR2A polymorphism and OS in an independent, larger trial dataset after adjusting for other potential prognostic factors.
2 METHODS
2.1 Study design and population
This retrospective, secondary analysis of thegerm line polymorphisms FCGR2A:H→R (rs1801274, cytosine→thymine) and FCGR3A:F→V (rs396991, cytosine→adenine) in wild-type KRAS patients used available DNA samples from the CCTG and the Australasian Gastro-Intestinal Trials Group (AGITG) CO.20 trial.21 Briefly, this was a multicenter, open-label, phase III randomized controlled trial; 750 chemotherapy-refractory metastatic colorectal cancer patients were randomized (1:1) to cetuximab and placebo vs cetuximab and brivanib alaninate, a dual inhibitor of vascular endothelial growth factor receptor and fibroblast growth factor receptor tyrosine kinase.22 Three months after study initiation, the protocol was amended to enroll only patients with wild-type KRAS, given new information regarding the lack of benefit of anti-EGFR monoclonal antibodies in KRAS mutant colorectal cancer.2 Twenty-one patients with mutated KRAS and four patients with indeterminable KRAS status were enrolled prior to the amendment. Our analysis was conducted on known wild-type KRAS patients only. Patients in both arms received cetuximab intravenously at an initial loading dose of 400 mg/m2 over 120 minutes, followed by a weekly maintenance infusion of 250 mg/m2 over 60 minutes. Patients randomly assigned to the combination arm also received oral brivanib at 800 mg on a daily schedule. No significant difference in the primary outcome of OS was observed (8.8 months vs 8.1 months in the brivanib and the placebo groups, respectively, HR: 0.88, 95% CI: 0.74-1.03; P = 0.12), despite a strong progression-free survival (PFS) benefit favoring the experimental arm with both cetuximab and brivanib (HR: 0.72; 95% CI: 0.62-0.84; P < 0.001).21
2.2 Outcomes
The primary objective of the current analysis was to evaluate the previously described association between FCGR2A polymorphism and OS in cetuximab-treated patients. Exploratory objectives included the association of FCGR2A polymorphism and PFS and the associations of FCGR3A polymorphism and OS and PFS. OS was defined as the time from random assignment until death from any cause. PFS was defined as the time from random assignment until the first observation of disease progression or death from any cause. The CCTG trial database was used for all analyses. REMARK guidelines were followed.23 All outcomes were planned prior to analysis initiation.
2.3 DNA extraction and genotyping method
Whole blood samples from local sites were archived at the CCTG central tissue bank (Queen's University, Kingston, Canada). DNA was extracted using the Qiagen whole blood DNA Kit. DNA quantity (spectrophotometry) and quality (polymerase chain reactions) were checked.DNA was independently genotyped blindly in the laboratory of G. Liu using TaqMan assays ordered from Thermo Fisher scientific company (Waltham, MA, USA). Assay IDs were C_9077561_20 for rs1801274, and C_25815666_10 for rs396991, with a subset of 30 confirmed also by Sanger sequencing.20 Results were checked using Hardy-Weinberg equilibrium testing.24 KRAS status was previously tested as a requirement for the CO.20 trial after the amendment.21
2.4 Statistical analysis
For the primary outcome analysis,FCGR2A polymorphism was compared with OS for all patients with genotyping results. Analyses were performed separately under the dominant, codominant, and additive genetic inheritance model assumptions; genotype was analyzed as a numerical variable for the additive model (number of copies of the minor allele), and as a categorical variable for the dominant and codominant models. The dominant genetic inheritance model was utilized for the primary analysis, consistent with our previous analysis.20 A formal power calculation for replication of our previous results under the dominant genetic inheritance model, assuming Hardy-Weinberg equilibrium, found empirical power of 97.5% for the univariate HR and of 99.7% for the adjusted HR, at a significance level of 0.5%.25 For the exploratory analyses, FCGR2A polymorphism was compared with PFS, and FCGR3A polymorphism was compared with OS and PFS. Interaction between FCGR2A and FCGR3A polymorphisms was explored by comparing outcomes of patients with both H/H FCGR2A and F/F FCGR3A polymorphisms to those with the R/R and V/V polymorphisms and to the rest of the study cohort, as a recent meta-analysis found the F/F FCGR3A genotype was associated with better OS.12 Additional exploratory analyses assessed potential relationships of FCGR2A and FCGR3A polymorphisms with toxicity. OS and PFS were assessed using Kaplan-Meier curves, log-rank tests (univariable analyses), and Cox proportional hazard models in multivariable analyses, adjusting for clinically relevant factors identified in the original trial analysis, including age, gender, side of primary tumor, stage, grade, number of metastatic sites, serum lactate dehydrogenase concentration, and treatment arm.21 Genomewide association studies (GWAS) generated a principal component analysis that was used to adjust for population stratification in the multivariable analyses. Tests for the assumption of proportional hazards were performed. R software version 3.3.1 (R Foundation for Statistical Computing, Vienna, Austria) was used for all analyses.
3 RESULTS
3.1 Patient and genotyping characteristics
The CO.20 trial included 725 patients who were known to be wild-type KRAS, of whom 595 (82%) had provided consent for genotyping and had available DNA extracted from an EDTA-treated whole blood sample. A total of 592 (99.4%) were successfully genotyped for FCGR2A and 594 (99.8%) for FCGR3A. The consort diagram of samples is shown in Figure 1. Demographic and disease variables are summarized in Table 1. A total of 165 patients (28%) had the H/H FCGR2A genotype, 299 (50%) had H/R, and 128 (22%) had R/R A total of 232 (39%) patients had the F/F FCGR3A genotype, 275 (46%) had F/V, and 87 (15%) had V/V. Both polymorphisms were in Hardy-Weinberg equilibrium: P = 0.73 (FCGR2A) and P = 0.71 (FCGR3A). Linkage disequilibrium between FCGR2A and FCGR3A was low (r2 = 0.03, D′ = 0.25). Patient characteristics were not significantly associated with FCGR2A polymorphisms (Table 1) and were also comparable between the subsets of patients with or without genotyping data (Table S1). Distribution of FCGR2A and FCGR3A genotypes was comparable in the brivanib and control study arms (Table S2). There was 100% concordance between the TaqMan assays and the Sanger sequencing.

| Characteristics | Genotyped patients (n = 592) | FCGR2A H/H (n = 165) | FCGR2A H/R (n = 299) | FCGR2A R/R (n = 128) | P-value |
|---|---|---|---|---|---|
| Mean age, y (SD) | 62.8 (10.7) | 62.4 (11.1) | 63 (10) | 62.9 (12.1) | 0.64 |
| Male gender | 392 (66%) | 115 (70%) | 190 (64%) | 87 (68%) | 0.37 |
| Side of primary cancer | |||||
| Left | 261 (44%) | 74 (45%) | 134 (45%) | 53 (41%) | 0.25 |
| Right | 125 (21%) | 26 (16%) | 66 (22%) | 33 (26%) | |
| Rectal | 206 (35%) | 65 (39%) | 99 (33%) | 42 (33%) | |
| Tumor stage at initial diagnosis | |||||
| I/II | 70 (12%) | 28 (18%) | 29 (10%) | 13 (11%) | 0.091 |
| III | 175 (31%) | 52 (32%) | 83 (29%) | 40 (33%) | |
| IV | 325 (57%) | 80 (50%) | 177 (61%) | 68 (56%) | |
| Missing | 22 | 5 | 10 | 7 | |
| Tumor grade | |||||
| I | 35 (6%) | 9 (6%) | 22 (8%) | 4 (4%) | 0.078 |
| II | 421 (78%) | 128 (83%) | 214 (77%) | 79 (72%) | |
| III | 86 (16%) | 17 (11%) | 42 (15%) | 27 (25%) | |
| Missing | 50 | 11 | 21 | 18 | |
| Number of metastatic sites | |||||
| ≤2 | 468 (79%) | 135 (82%) | 231 (77%) | 102 (80%) | 0.52 |
| >2 | 124 (21%) | 30 (18%) | 68 (23%) | 26 (20%) | |
| Number of previous lines of chemotherapy | |||||
| ≤2 | 22 (4%) | 6 (4%) | 9 (3%) | 7 (5%) | 0.44 |
| >2 | 570 (96%) | 159 (96%) | 290 (97%) | 121 (95%) | |
- SD, standard deviation.
3.2 Association between FCGR2A polymorphisms and OS
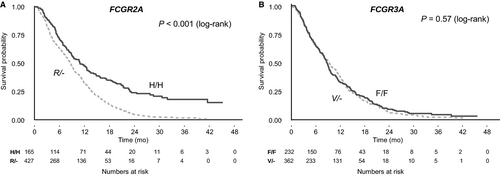
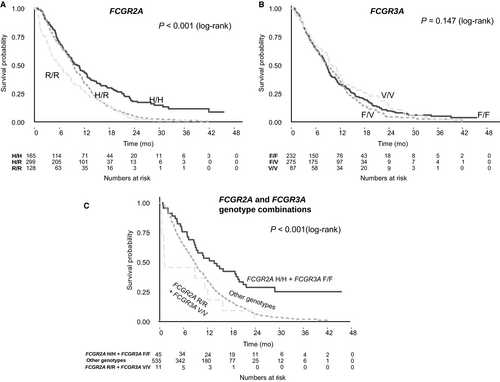
All Cox models were consistent with the assumption of proportional hazards. There was a statistically significant association between FCGR2A genotype and OS in the study cohort (Table 2). In the primary univariable analysis under the dominant inheritance model, the H/H genotype was associated with better OS than the R/- genotype (HR: 0.61, 95% CI: 0.5-0.74, P < 0.001, median absolute benefit 1.3 months; Figure 2). Similar results were obtained in the exploratory analyses under the additive model (OR: 0.71 per H allele, 95% CI: 0.63-0.8, P < 0.001) and the codominant model, with the H/H genotype associated with better OS compared with the R/R genotype (HR: 0.51, 95% CI: 0.4-0.65, P < 0.001, median absolute benefit 3.7 months), and intermediate outcomes for patients with the H/R genotype (HR: 0.65, 95% CI: 0.53-0.8, P < 0.001; Figure 3). Results remained significant in multivariable analysis under all models (Table 2). There was no interaction between FCGR2A polymorphism and OS according to treatment arm (P = 0.37).
| Genetic inheritance model | Genotype | Median survival (mo) | One-year survival (95% CI) | Two-year survival (95% CI) | aHR (95% CI) | P-value |
|---|---|---|---|---|---|---|
| Primary analysis | ||||||
| Dominant | H/H | 9.6 | 44% (37-52) | 18% (13-26) | 0.66 (0.54-0.81) | <0.001 |
| R/- | 8.3 | 32% (27-37) | 4% (2-8) | Reference | ||
| Exploratory analyses | ||||||
| Codominant | H/H | 9.6 | 44% (37-52) | 18% (13-26) | 0.53 (0.41-0.68) | <0.001 |
| H/R | 9.3 | 34% (29-40) | 5% (3-8) | 0.72 (0.58-0.89) | 0.003 | |
| R/R | 5.9 | 27% (21-36) | 3% (1-8) | Reference | ||
| Additive | H/H | 9.6 | 44% (37-52) | 18% (13-26) | Per increase in 1 wild-type | <0.001 |
| H/R | 9.3 | 34% (29-40) | 5% (3-8) | H allele: 0.72 (0.64-0.83) | ||
| R/R | 5.9 | 27% (21-36) | 3% (1-8) | |||
- Analyses are adjusted for clinically relevant factors identified in the original trial analysis.21 Genomewide association studies (GWAS), used for population stratification, were available for 566 of the 592 (95.6%) patients with genotyping results, and subset analyses of these 566 patients that included adjustment by principal components found virtually identical results.
- aHR, adjusted hazard ratio from Cox proportional hazards models; CI, confidence interval.
3.3 Exploratory associations between FCGR polymorphisms and clinical outcomes
There was a statistically significant association between FCGR2A genotype and PFS in the study cohort. In univariable analysis under the dominant inheritance model, the H/H genotype was associated with better PFS compared with the R/- genotype (HR: 0.80, 95% CI: 0.67-0.95, P = 0.01, median absolute benefit 1.6 months). Similar results were obtained under the additive inheritance model (OR: 0.80 per H allele, 95% CI: 0.71-0.9, P < 0.001) and under the codominant model, with the H/H genotype associated with better PFS than patients with the R/R genotype (HR: 0.63, 95% CI: 0.5-0.8, P < 0.001, median absolute benefit 3.3 months) and similar PFS compared with the H/R genotype (HR: 0.87, 95% CI: 0.71-1.05, P = 0.14; Figure S1). Results remained significant in multivariable analysis under all models (Table 3). There was no interaction between FCGR2A polymorphism and PFS according to treatment arm (P = 0.65).
| Polymorphism | Genotype | Median survival (mo) | Codominant model: aHR (95% CI), P-value | Additive model: aHR (95% CI), P-value | Dominant model: aHR (95% CI), P-value |
|---|---|---|---|---|---|
| Overall survival | |||||
| FCGR3A | F/F | 8.6 | 1.03 (0.78-1.35), 0.84 | Per increase in 1 wild-type F allele: 0.99 (0.87-1.12), 0.80 | F/F vs V/-: 0.93 (0.78-1.11), P = 0.44 |
| F/V | 8.8 | 0.91 (0.75-1.09), 0.30 | |||
| V/V | 9.5 | reference | |||
| FCGR2A and FCGR3A genotype combinations | H/H + F/Fa | 13.7 | 0.33 (0.16-0.68), 0.003 | Not applicable | Not applicable |
| Others | 8.7 | 0.52 (0.36-0.75), <0.001 | |||
| R/R + V/Va | 1.2 | Reference | |||
| Progression-free survival | |||||
| FCGR2A | H/H | 5.2 | 0.65 (0.51-0.83), <0.001 | Per increase in 1 wild-type H allele: 0.81 (0.72-0.92), <0.001 | H/H vs R/-: 0.80 (0.66-0.97), P = 0.02 |
| H/R | 3.7 | 0.87 (0.71-1.05), 0.15 | |||
| R/R | 1.9 | Reference | |||
| FCGR3A | F/F | 3.7 | 0.99 (0.76-1.28), 0.92 | Per increase in 1 wild-type F allele: 0.97 (0.86-1.1), 0.64 | F/F vs V/-: 0.93 (0.78-1.1), P = 0.39 |
| F/V | 3.6 | 0.91 (0.76-1.09), 0.31 | |||
| V/V | 5.1 | Reference | |||
| FCGR2A and FCGR3A genotype combinations | H/H + F/Fa | 5.5 | 0.45 (0.22-0.92), 0.03 | Not applicable | Not applicable |
| Others | 3.6 | 0.79 (0.58-1.09), 0.15 | |||
| R/R + V/Va | 1.0 | Reference | |||
- Analyses are adjusted for clinically relevant factors identified in the original trial analysis.21 Genomewide association studies (GWAS), used for population stratification, were available for 566 of the 592 (95.6%) patients with genotyping results, and subset analyses of these 566 patients that included adjustment by principal components found virtually identical results.
- aHR, adjusted hazard ratio from Cox proportional hazards models; CI, confidence interval.
- Double wild-type genotype (wild type for both FCGR2A and FCGR3A).
- Double-homozygous variant genotype (homozygous variant for both FCGR2A and FCGR3A).
No association was observed between FCGR3A polymorphism and OS (Table 3, Figures 2 and 3) or PFS (Table 3, Figure S1). However, when combinations of genotypes of FCGR2A and FCGR3A polymorphisms were compared with OS and PFS both in univariable analysis and adjusted for key known prognostic factors, patients with both the H/H FCGR2A and the F/F FCGR3A double wild-type genotypes had significantly better OS compared to those with the R/R and V/V polymorphisms (HR: 0.33, 95% CI: 0.16-0.68, P = 0.003; median absolute benefit, 12.5 months) and those with any other genotype (Table 3, Figure 3).
3.4 Exploratory association between FCGR polymorphisms and toxicity
There was no association between FCGR2A (P = 0.13) or FCGR3A (P = 0.64) and any grade 3 or greater treatment toxicity in multivariable analysis. A pre-specified analysis of the interaction between FCGR2A and FCGR3A polymorphisms and skin rash also found no statistically significant interaction, P = 0.20 and P = 0.71, respectively. Exploratory analyses further found virtually identical relationships in subset analyses by trial arm.
4 DISCUSSION
We have successfully replicated our previous results demonstrating an association between FCGR2A genotype and survival in wild-type KRAS colorectal cancer patients treated with cetuximab. H/H genotype was associated with significantly longer OS than the R/- genotype under the dominant genetic model assumptions. Additional exploratory analyses under the additive and codominant genetic inheritance models found intermediate results for patients with the heterozygous genotype; this finding expands on the prior study which had too small a sample size to adequately assess these models. The similar and consistently significant results under the additive and codominant genetic inheritance models imply that the clinical benefit for colorectal cancer patients treated with cetuximab could be additive in nature, with an improvement of approximately ~30% in survival benefit for every H allele of FCGR2A.
The large sample size was also used to explore the joint effect of both FCGR2A and FCGR3A polymorphisms together; we found that the best outcomes were in patients with the double wild-type genotypes and the worst were found in the 2% of patients with the double-homozygous variant genotypes, with a threefold difference in OS (absolute median difference 12.5 months) after adjustment for other prognostic factors, although confidence intervals were wide. As the presence of both homozygous variants is a rare occurrence, the additional clinical utility of this finding is limited. However, it provides further insight into the mechanistic role of FCGR polymorphisms in clinical outcomes of cetuximab-treated, metastatic colorectal cancer, namely, the role of FCGR3A polymorphism in modifying the primary relationship between FCGR2A and clinical outcomes.
Regardless, the primary clinical implication of this study relates to the common variant allele frequency of the FCGR2A polymorphism26: OS was twice longer for patients with the FCGR2A H/H polymorphism than those with the R/R genotype, corresponding to median absolute difference of 3.7 months in the patient population now known to benefit from cetuximab, namely KRAS wild-type patients. Of additional importance is that genotypes associated with better OS were not associated with higher rates of grade 3 or greater toxicity, nor were they related to skin toxicity. This is likely due to the different mechanisms involved: Toxicity is thought to be mediated through competitive inhibition of the EGFR receptor,27, 28 a different mechanism of action from Fc-γ receptor-regulated ADCC.3, 4 No interaction was found between treatment arm and the analyzed polymorphisms, which was to be expected as both treatment arms included the same cetuximab regimen, and brivanib efficacy was not expected to be affected by FCGR polymorphisms.
Although our findings replicate the results of several prior publications8, 14, 15 and of our previous analysis,20 others have reported different results. Notably, a recent consortium analysis of 660 cetuximab-treated patients with wild-type KRAS metastatic colorectal carcinoma did not find any association between FCGR2A polymorphisms and clinical outcome.16 However, the study population was heterogeneous in regard to the studied intervention, with a minority of the patients treated with single-agent cetuximab, and most patients treated with irinotecan-based regimens that included cetuximab.16 Also, only approximately a third of the patients had available germ line DNA from blood samples, with the rest of the patients having putatively normal DNA extracted from formalin-fixed paraffin-embedded (FFPE) slides,16 an approach that has been demonstrated to be unreliable for at least some genes.30 Both of our studies were conducted in relatively homogenous and prospective patient populations with germ line DNA genotyped from blood samples, and the results replicated.23
While the FCGR2A wild-type genotype was associated with better OS and PFS, this study's results were of greater magnitude for OS, consistent with our previous findings in CO.17,20 and with several large studies of anti-EGFR mAbs for wild-type KRAS colorectal cancer, such as the FIRE-3 and the PEAK trials.31, 32 Although OS can be confounded by post-trial treatments, it is less likely to be affected by pre-trial confounders, given the randomized nature of these trials. An alternative explanation might be a joint effect between FCGR polymorphisms and anti-EGFR mAbs, untested in the aforementioned trials and identified in this trial, causing failure of surrogacy of PFS for OS.
Similar interactions between FCGR2A polymorphism and clinical outcomes have been reported for other mAbs and malignancies. Patients with germ line H/H polymorphism treated with the anti-human epidermal growth factor receptor 2 (HER2) mAb trastuzumab for early HER2-positive breast cancer were more likely to achieve pathological response while those with metastatic disease had better objective response rate and PFS.33, 34 The H/H polymorphism of FCGR2A was also associated with better response to anti-TNF mAbs in patients with rheumatoid arthritis.36, 37 This evidence further supports a genetic role in determining cetuximab treatment outcomes.
Limitations of this study include an inability to genotype the entire sample as the blood sample component of the CO.20 trial was voluntary. However, the genotyped subgroup represented 79% of the entire trial population, with similar clinical and demographic characteristics (Table S1). Wild-type KRAS status was defined based on the assessment of lack of mutations seen in exon 2; the effect of other, significantly rarer RAS and BRAF mutations on this polymorphism-outcome association, was not assessed in this analysis.40 EGFR polymorphisms previously reported to be associated with cetuximab efficacy and toxicity were also not assessed.41, 42 Finally, although we could perform joint analyses of combinations of FCGR2A and FCGR3A genotypes, we still had an insufficient sample size to perform a formal interaction analysis.
In conclusion, this analysis confirms that FCGR2A H/H polymorphism in patients with wild-type KRAS metastatic colorectal cancer treated with cetuximab is associated with significantly longer OS without affecting toxicity profiles. This association may be modified by the FCGR3A polymorphism, as patients with the double wild-type genotypes of H/H and F/F genotypes had the best clinical outcomes. This replication in a large, separate dataset provides evidence to evaluate prospectively the utility of FCGR polymorphisms as biomarkers for clinical management in this patient population.
CONFLICT OF INTEREST
The authors declare no potential conflict of interests.




