Crosstalk between metabolic reprogramming and epigenetics in cancer: updates on mechanisms and therapeutic opportunities
Tongxin Ge and Xiang Gu contributed equally.
Abstract
Reversible, spatial, and temporal regulation of metabolic reprogramming and epigenetic homeostasis are prominent hallmarks of carcinogenesis. Cancer cells reprogram their metabolism to meet the high bioenergetic and biosynthetic demands for vigorous proliferation. Epigenetic dysregulation is a common feature of human cancers, which contributes to tumorigenesis and maintenance of the malignant phenotypes by regulating gene expression. The epigenome is sensitive to metabolic changes. Metabolism produces various metabolites that are substrates, cofactors, or inhibitors of epigenetic enzymes. Alterations in metabolic pathways and fluctuations in intermediate metabolites convey information regarding the intracellular metabolic status into the nucleus by modulating the activity of epigenetic enzymes and thus remodeling the epigenetic landscape, inducing transcriptional responses to heterogeneous metabolic requirements. Cancer metabolism is regulated by epigenetic machinery at both transcriptional and post-transcriptional levels. Epigenetic modifiers, chromatin remodelers and non-coding RNAs are integral contributors to the regulatory networks involved in cancer metabolism, facilitating malignant transformation. However, the significance of the close connection between metabolism and epigenetics in the context of cancer has not been fully deciphered. Thus, it will be constructive to summarize and update the emerging new evidence supporting this bidirectional crosstalk and deeply assess how the crosstalk between metabolic reprogramming and epigenetic abnormalities could be exploited to optimize treatment paradigms and establish new therapeutic options. In this review, we summarize the central mechanisms by which epigenetics and metabolism reciprocally modulate each other in cancer and elaborate upon and update the major contributions of the interplays between epigenetic aberrations and metabolic rewiring to cancer initiation and development. Finally, we highlight the potential therapeutic opportunities for hematological malignancies and solid tumors by targeting this epigenetic-metabolic circuit. In summary, we endeavored to depict the current understanding of the coordination between these fundamental abnormalities more comprehensively and provide new perspectives for utilizing metabolic and epigenetic targets for cancer treatment.
Abbreviations
-
- PPP
-
- Pentose phosphate pathway
-
- PDA
-
- Pancreatic ductal adenocarcinoma
-
- ncRNAs
-
- non-coding RNAs
-
- CpG
-
- cytosine-guanine
-
- DNMT
-
- DNA methyltransferase
-
- TET
-
- Ten-eleven translocation family proteins
-
- HAT
-
- Histone acetyltransferase
-
- HDAC
-
- Histone deacetylase
-
- SIRT
-
- Sirtuin
-
- KMT
-
- Histone lysine methyltransferase
-
- SAM
-
- S-adenosyl methionine
-
- KDM
-
- Histone lysine demethylase
-
- LSD
-
- Lysine-specific demethylase
-
- FAD
-
- Flavin adenine dinucleotide
-
- JHDM
-
- Jumonji C domain-containing histone demethylase
-
- α-KG
-
- α-ketoglutarate
-
- CRC
-
- Chromatin remodeling complex
-
- ncRNA
-
- Non-coding RNA
-
- miRNA
-
- MicroRNA
-
- lncRNAs
-
- Long non-coding RNA
-
- circRNA
-
- Circular RNA
-
- NADPH
-
- Reduced nicotinamide adenine dinucleotide phosphate
-
- TCA cycle
-
- Tricarboxylic acid cycle
-
- SAH
-
- S-adenosyl homocysteine
-
- NAD+
-
- Nicotinamide adenine dinucleotide
-
- 2-HG
-
- 2-hydroxyglutarate
-
- Acetyl-CoA
-
- Acetyl-coenzyme A
-
- ACL
-
- ATP-citrate lyase
-
- ACSS2
-
- Acetyl-CoA synthetase 2
-
- PDC
-
- Pyruvate dehydrogenase complex
-
- LKB1
-
- Liver kinase B1
-
- SHMT2
-
- Serine hydroxymethyltransferase 2
-
- PHGDH
-
- Phosphoglycerate dehydrogenase
-
- NEPC
-
- Small cell/neuroendocrine prostate cancer
-
- PKCλ/ι
-
- Protein kinase C λ/ι
-
- MAT2A
-
- Methionine adenosyltransferase 2A
-
- NNMT
-
- Nicotinamide N-methyltransferase
-
- 1MNA
-
- 1-methylnicotinamide
-
- YTHDF2
-
- YTH N6-methyladenosine RNA binding protein 2
-
- m6A
-
- N6-methyladenosine
-
- SCC
-
- Squamous cell carcinoma
-
- OAADPR
-
- 2′-O-acyl-ADP ribose
-
- FAO
-
- Fatty acid oxidation
-
- NAM
-
- Nicotinamide
-
- NAMPT
-
- Nicotinamide phosphoribosyltransferase
-
- NMNAT-1
-
- NMN adenylyltransferase 1
-
- IDH
-
- Isocitrate dehydrogenase
-
- LDHA
-
- Lactate dehydrogenase A
-
- SDH
-
- Succinate dehydrogenase
-
- FH
-
- Fumarate hydratase
-
- GIST
-
- Gastrointestinal stromal tumor
-
- AML
-
- Acute myeloid leukemia
-
- EZH2
-
- Enhancer of zeste homolog 2
-
- BCAT1
-
- Branched-chain amino acid transaminase 1
-
- HK2
-
- Hexokinase 2
-
- G6PD
-
- Glucose-6-phosphate dehydrogenase
-
- ROS
-
- Reactive oxygen species
-
- SETD2
-
- SET domain-containing 2
-
- G9A
-
- Euchromatic histone-lysine N-methyltransferase 2
-
- PSAT1
-
- Phosphoserine aminotransferase 1
-
- PSPH
-
- Phosphoserine phosphatase
-
- HCC
-
- Hepatocellular carcinoma
-
- ARID1A
-
- AT-rich interacting domain-containing protein 1A
-
- GLS
-
- Glutaminase
-
- GSH
-
- Reduced glutathione
-
- SMARCA4
-
- SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4
-
- RISC
-
- RNA-induced silencing complex
-
- ENO1
-
- Enolase 1
-
- PKM2
-
- Pyruvate kinase isoform M2
-
- CPT1
-
- Carnitine palmitoyl transferase 1
-
- PFKFB3
-
- 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
-
- LUAD
-
- Lung adenocarcinoma
-
- GOT1
-
- Glutamic-oxaloacetic transaminase
-
- GLUT1
-
- Glucose transporter type 1
-
- NPC
-
- Nasopharyngeal carcinoma
-
- PFK2
-
- 6-phosphofructo-2-kinase
-
- MTHFD2
-
- Methylenetetrahydrofolate dehydrogenase 2
-
- ccRCC
-
- Clear cell renal cell carcinoma
-
- FTO
-
- Fat mass and obesity-associated protein
-
- LDHB
-
- Lactate dehydrogenase B
-
- METTL3
-
- Methyltransferase-like 3
-
- OXPHOS
-
- Oxidative phosphorylation
-
- ALKBH5
-
- AlkB homolog 5 RNA demethylase
-
- RCC
-
- Renal cell carcinoma
-
- METTL14
-
- Methyltransferase-like 14
-
- m5C
-
- 5-methylcytosine
-
- SAMTOR
-
- SAM sensor upstream of mTORC1
1 BACKGROUND
Cellular metabolic reprogramming is a core hallmark of cancer [1, 2]. A large body of researches have tried to elucidate the direct effects of metabolism on tumor growth, proliferation, and metastasis. Highly proliferating cancer cells require numerous building blocks for active biosynthesis and an abundant energy supply. To meet the requirements for growth and survival, cancer cells experience significant metabolic alterations, such as upregulated glycolysis and enhanced glutamine catabolism. Oncogenic reprogramming of cellular metabolism is a downstream event of mutant oncogenes or tumor suppressors, dysregulated signal transduction pathways, and perturbed microenvironmental nutrient availability [3-6]. Emerging researches suggest that metabolism is not merely a passive participant of tumorigenesis; it can serve as signaling molecules and globally control gene expression. Another general mechanism by which metabolism can modulate cellular activities has been proposed. Cellular metabolism provides a pool of intermediate metabolites acting as substrates, cofactors, agonists, or antagonists of chromatin-modifying enzymes. Significant changes in the metabolic pool accompany the reprogramming of metabolism. Thus, it is reasonable to speculate that fluctuations in these metabolites could regulate the state and function of cells through epigenetic mechanisms. The hyperactive pentose phosphate pathway (PPP) promotes global epigenomic reprogramming and drives the evolution of distant metastasis in pancreatic ductal adenocarcinoma (PDA), providing robust evidence for this hypothesis [7].
The term “epigenetics” was defined as a “stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence” [2, 8]. Beyond oncogenic mutations, four classic epigenetic mechanisms, DNA methylation, histone modifications, chromatin remodeling, and non-coding RNAs (ncRNAs), dynamically influence various chromatin-related processes, such as gene transcription, DNA repair, and replication. The basic unit of chromatin is the nucleosome, which is assembled from a histone octamer consisting of H2A, H2B, H3, and H4, with 147 base pairs of DNA wrapped around the octamer [9]. Alterations in chromatin structure caused by epigenetic modifications and chromatin remodelers can change the transcriptional accessibility of regional DNA sequences, thus profoundly influencing gene expression. In human cancers, epigenetic modification profiles and ncRNA expression patterns often change globally [10-14]. Compelling evidence highlights that epigenetic reprogramming is crucial for the acquisition and maintenance of hallmark capabilities in cancer, including unlocking phenotypic plasticity and deregulating cellular metabolism [2, 15-19].
Some studies have revealed that the interplay between epigenetics and metabolic reprogramming endows tumor cells with the capability to adapt to ever-changing conditions during tumorigenesis. Most recently, many discoveries have been made. These findings will be discussed in detail later to provide more supporting evidence for this hypothesis. Additionally, with advances in the fields of cancer metabolism and epigenetics, several intriguing new themes have emerged. One key question is how metabolism tunes transcription through non-canonical histone modifications like lactylation and succinylation. A second important question is whether a close interaction exists between metabolism and RNA epigenetics. Covering these themes will significantly deepen our understanding of this topic and provide fundamental insights into tumor biology. However, there are still some limitations existing in current studies. First, the causal link between the metabolic-epigenetic loop and phenotypic outcomes in cancer has not been rigorously proven. That is to say, whether all these outcomes observed are directly caused by metabolically driven changes in epigenetic modifications needs further validation. Newly developed epigenome editing may enable us to confirm which chromatin marks have causal roles in determining tumor behaviors [20]. Second, metabolic and epigenetic heterogeneities within tumors are currently rarely taken into account. High-throughput techniques, including spatial omics and single-cell omics, may answer the question of how heterogenous metabolic and epigenetic patterns interweave with each other to amplify intra-tumoral phenotypic diversity [21].
Cancer metabolism and epigenetics are both attractive therapeutic targets for cancer therapy, which is not surprising, given their important roles in cancer. Unfortunately, successful clinical applications of drugs targeting metabolism are rare. The efficacy of epigenetic drugs has been confined to hematological malignancies, and they are almost ineffective in solid tumors. This indicates the need to identify true metabolic or epigenetic vulnerabilities and develop new drug combinations. The robust association between metabolism and epigenetics has been revealed. It is thus rational to propose some potential treatment strategies targeting these communications (Figure 1).
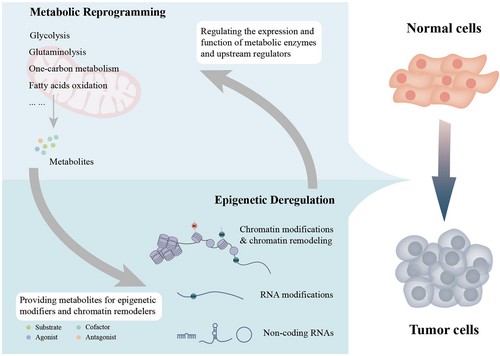
2 REPROGRAMMED CELLULAR METABOLISM IN CANCER
The most classic example of metabolic reprogramming in cancer is the Warburg effect, also known as aerobic glycolysis. Cancer cells tend to convert pyruvate, the end product of glycolysis, into lactate rather than directing it into mitochondrial metabolism despite the intact function of oxidative phosphorylation (OXPHOS). This may be caused by the increased demand of cancer cells for macromolecule biosynthesis compared with energy production. The intermediate products of glycolysis can be diverted into biosynthetic programs such as serine metabolism, hexosamine pathway, and PPP [22, 23]. These metabolic branches, often deregulated, provide reduced nicotinamide adenine dinucleotide phosphate (NADPH) for reductive biosynthesis and combating oxidative stress and S-adenosyl methionine (SAM) for methylation reactions and building blocks for proteins and nucleic acids [24, 25]. In addition, cancer cells can utilize intermediates of the tricarboxylic acid (TCA) cycle for de novo fatty acid and non-essential amino acid synthesis. Researchers reported that cancer cells might be addicted to glutamine and glucose [26, 27]. Glutamine is involved in the synthesis of essential amino acids, purine bases, and pyrimidine bases. Further, glutamine can also be metabolized into α-ketoglutarate (α-KG) to replenish the TCA cycle in the mitochondria [28]. Beyond that, lipid metabolism also undergoes reprogramming in cancer. Cancer cells have active fatty acid and cholesterol synthesis that make up the membrane and form signaling molecules. Fatty acid oxidation is an important energy source for rapidly proliferating cancer cells [29, 30]. Altogether, metabolic reprogramming dramatically impacts many biological properties of cancer cells, such as fueling proliferation and growth and promoting invasion and distant metastasis [31, 32].
3 EPIGENETIC MECHANISMS IN CANCER
Epigenetic changes, including DNA methylation, histone modifications, chromatin remodeling, and ncRNAs, are closely related to cancer development and malignant progression. Here, we provide an overview of the basic principles of these epigenetic processes.
3.1 DNA methylation
DNA methylation refers to the enzymatic addition of a methyl group to a cytosine 5-carbon, which forms 5-methylcytosine (5mC). It occurs mainly at scattered cytosine-guanine (CpG) dinucleotide sites and some CpG islands, which are CpG-rich sequences [33]. Nevertheless, CpG sites in CpG islands that overlap with the promoter regions of approximately two-thirds of human genes are commonly unmethylated to maintain a permissive chromatin state for transcription [34]. In cancer, DNA methylation patterns are extensively reshaped with global hypomethylation but with regional hypermethylation of CpG islands in promoters of tumor suppressor genes [33, 35]. DNA methyltransferases (DNMTs) utilize SAM as the methyl group donor and are responsible for the deposition of methyl groups on C5 of cytosines [36]. DNMTs include two major categories: maintenance methyltransferase DNMT1 and de novo methyltransferases DNMT3A and DNMT3B. Ten-eleven translocation (TETs) family proteins, including TET1, TET2, and TET3, have been demonstrated to be mammalian DNA hydroxylases for active DNA demethylation. TETs require oxygen and α-KG as substrates and ferrous iron as cofactors to mediate demethylation reactions [37]. Specifically, 5mC is oxidized stepwise into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) during this process, followed by replication-dependent dilution or base excision repair [38]. Of note, 5hmC represents both a demethylation intermediate and a stable epigenetic mark. Its abundance could reflect the function and activity of TETs [39, 40].
In cancer cells, global DNA hypomethylation revealed by genome-wide analyses is the most prominent and earliest identified DNA methylation abnormality [41]. DNA hypomethylation, accompanied by the activation of transcription, repeats, transposable elements, and oncogenes, may contribute to increases in aneuploidies and genomic instability, which are hallmarks of cancer [42]. Furthermore, aberrant hypermethylation of CpG islands in the 5′ promoter regions of tumor suppressor genes in cancer cells can lock them into inactive states, silencing their expression. For example, RB, a well-known tumor suppressor gene, was discovered to be downregulated by promoter CpG-islands hypermethylation and promote oncogenesis [43, 44]. Such aberrant DNA methylation patterns were also observed in tumor suppressor genes like CDKN2A, MLH1, and CDH1 [45-47].
3.2 Histone modifications
Each histone possesses a highly flexible N-terminal tail enriched with lysine and arginine residues that can be extensively modified [48]. Covalent histone modifications include acetylation, methylation, acylation (e.g., lactylation, succinylation, and crotonylation), phosphorylation, SUMOylation, and citrullination. Some histone modifications can alter interactions between histones and DNA or can be recognized by specific binding proteins to impact chromatin compaction and regulate transcription processes [49, 50].
Histone acetylation can promote a more open chromatin state and increase chromatin accessibility for gene expression. Histone acetylation is dynamically established by histone acetyltransferases (HATs) and is removed by histone deacetylases (HDACs). There are three major groups of HATs: the GNAT family, the MYST family, and the orphan family. HATs transfer acetyl groups from acetyl-coenzyme A (acetyl-CoA) to histone lysine residues [51]. Four classes of HDACs were identified: class I (HDAC1-3 and HDAC8), class II (HDAC4-7 and HDAC9-10), class IV (HDAC11), and class III (Sirtuin/SIRT1-7). SIRTs require nicotinamide adenine dinucleotide (NAD+) as the cofactor [51, 52]. Some HDACs can also deacetylate nonhistone proteins [52].
Histone methylation occurs in the side chains of lysine, arginine, and histidine residues. Histone lysine methyltransferases (KMTs) can specifically transfer one, two, or three methyl groups from SAM to specific histone lysine residues to generate mono-, di-, or tri-methylated (me1/2/3) histone [53]. There are two kinds of histone demethylases (KDMs). The family of amine oxidases (LSDs) utilizes flavin adenine dinucleotide (FAD) as a cofactor and is limited to demethylating mono- and di-methylated lysine. Jumonji C (JmjC) domain-containing histone demethylases (JHDMs) utilize ferrous iron and α-KG and demethylate tri-methylated lysine [54]. The functions of different histone methylations depend on the location and degree of methylation of lysine residues. Histone methylation plays an essential role in modulating transcription by changing the chromatin structure, recruiting chromatin remodeling factors, or guiding the binding of transcription factors [55].
In cancer cells, a genome-wide profile revealed the loss of mono-acetylation and tri-methylation of histone H4 at a global level [56]. The discovery was subsequently confirmed in skin cancer, and the study suggested that the alteration occurs at the early stage and accumulates during carcinogenesis. With growing evidence supporting this discovery in multiple cancers, it was accepted as a common feature of cancer cells. These losses primarily appeared at the acetylated K16 and tri-methylated K20 residues of histone H4 and were connected to the well-described DNA hypomethylation in cancer [56-58]. In addition, certain combinations of histone modification are associated with extensive CpG island hypermethylation in cancer cells, including H3K9 methylation, H3K27 tri-methylation, loss of H3K4 tri-methylation, and deacetylation of histones H3 and H4 [59, 60]. Histone modifications promote tumor pathogenesis and evolution through transcriptional regulation that activates oncogene expression and represses tumor suppressor gene expression. For example, the enhancer of zeste homolog 2 (EZH2) binds to the promoter region of P21, a crucial tumor suppressor gene, and regulates its H3K27me3 modification, which promotes proliferation and tumorigenesis in gastric cancer [61].
3.3 Chromatin remodeling
Chromatin structure is dynamically regulated by DNA and histone modifiers and ATP-dependent chromatin remodeling complexes (CRCs). CRCs contain four different families: switch/sucrose non-fermentable (SWI/SNF), imitation switch (ISWI), chromodomain-helicase DNA-binding (CHD), and inositol-requiring mutant 80 (INO80). CRCs can change the packaging state of chromatin, specialize in chromatin regions, and regulate chromatin accessibility through sliding, ejecting, or reorganizing nucleosomes [62, 63]. Components of the SWI/SNF complex are frequently and extensively mutated in various types of cancer; however, the mechanisms of CRCs mutations in tumorigenesis remain unclear [64].
The SWI/SNF family, composed of 8 to 14 subunits, was initially extracted from Saccharomyces cerevisiae. Eukaryotes usually employ two SWI/SNF family complexes with two relevant catalytic subunits. The family slides and ejects nucleosomes in various processes at many loci but is incapable of chromatin assembly [65]. The ISWI family comprises 2 to 4 subunits. Among the ISWI family, dNURF, dCHRAC and dACF complexes were initially extracted from Drosophila melanogaster and hWICH or hNoRC was subsequently recognized. Eukaryotes develop diverse ISWI family complexes by combining one or two catalytic subunits with specialized proteins [66]. Most ISWI family complexes, including ACF and CHRAC, promote chromatin assembly and transcriptional repression by improving nucleosome spacing [62]. The CHD family, among which Mi-2 combines 1 to 10 subunits, was initially extracted from Xenopus laevis [67]. Some CHD family complexes promote transcription by sliding or ejecting nucleosomes, whereas others repress transcription, including the vertebrate Mi-2/NuRD complex. The variability in CHD family complexes may result in chromodomain diversity [68]. The INO80 family, composed of more than 10 subunits, was first extracted from Saccharomyces cerevisiae. INO80 participates in DNA repair and transcriptional activation [69]. Notably, SWR1-related complexes in the INO80 family reorganize nucleosomes by replacing canonical H2A-H2B dimers with H2A.Z-H2B dimers [70].
So far, the studies of chromatin remodeling in cancer have focused on SWI/SNF family. The sequencing of cancer genomes revealed high-frequency mutations in various SWI/SNF family members in several hematological and solid malignancies, including SNF5, BRG1, MTA1 and ARID1A [71-75]. These members act as tumor suppressors, the mutations of which contribute to the development and maintenance of cancer. The mutations of these chromatin remodelers provided opportunities to change chromatin accessibility and protein complex topology, yielding oncogenic outcomes. Mutations in the SMARCB1 gene promote tumorigenesis in malignant rhabdoid tumors by preventing SWI/SNF complex interaction with typical enhancers and promoting remaining SWI/SNF subunits to induce gene expression at super-enhancers [76]. In addition, the SWI/SNF family complexes interact with transcription factors by multiple lineage-specific subunits to regulate differentiation. They also potentiate malignancy by damaging the balance between differentiation and self-renewal. Moreover, SWI/SNF family complexes participate in cell motility, cell-cycle progression, and nuclear hormone signaling [75].
3.4 Non-coding RNAs
ncRNAs are functional transcripts driven by non-protein-coding genomes. Among the ncRNA family, microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) are relatively well studied in cancer. They functionally interact with each other and form a sophisticated regulatory network, finely regulating all fundamental biological processes in cells [77, 78].
MiRNAs are small ncRNAs containing about 22 nucleotides, biogenesis taking place through a multi-step process involving the RNase III enzymes, Drosha and Dicer [79]. They inhibit post-transcriptional gene expression by regulating mRNA translation into proteins and are estimated to mediate the translation of over 60% of protein-coding genes. The inhibition is completed through mRNA degradation and the suppression of translation initiation [80]. MiRNAs participate in multiple biological processes, including development, proliferation, differentiation, and apoptosis. Some miRNAs mediate specific individual targets, while others function as major process controllers, simultaneously regulating multiple gene expressions [81].
LncRNAs, comprising the largest portion of the non-coding transcriptome, are a heterogeneous group encompassing transcripts longer than 200 nucleotides and without protein-coding capacity [82]. Although lncRNAs were considered to lack open reading frames or conserved codons in transcripts, the recent investigation suggested that some transcripts may produce small peptides [83, 84]. Compared to protein-coding genes, lncRNAs are commonly expressed at a lower level but display more cell type-specific expression patterns. Functions of lncRNAs are more complicated and varied than that of miRNAs, including transcriptional regulation, mRNA processing, and post-transcriptional control [77].
CircRNAs are characterized by the covalent link of the 3′ and 5′ ends in forming single-stranded continuous loop structures, reshaping RNA structure cognition [85]. They are more stable than liner ncRNAs, owing to the lack of exposed ends that are inclined to nucleolytic degradation and specific RNA folding. In addition, they are evolutionary conserved and abundant in eukaryotes [86]. Splicing and circularization of exons or introns are considered the initial genesis events of circRNAs. CircRNAs exert critical biological functions by serving as sponges to inhibit miRNAs, mediating protein functions or encoding peptides [87].
Growing evidence has revealed that the aberrant expression of ncRNAs is one of the hallmark features of cancers, and distinct ncRNA expression profiles exhibited between tumor cells and normal cells play a vital role in tumor progression and metastasis [88, 89]. Cancer-associated miRNAs are commonly categorized into tumor suppressor miRNAs and oncogenic miRNAs. Well-established tumor suppressor miRNAs involve miR-145, miR-34a, and the let-7 family and well-characterized oncogenic miRNAs include miR-21 and miR-155 [90]. Notably, some miRNAs exert dual functions. For example, miR-200c constrains epithelial-to-mesenchymal transition (EMT) to inhibit metastasis initiation in cancer; however, it promotes distant metastasis in late-stage cancers [91-93]. Notably, miRNAs can inhibit cell proliferation by targeting cell cycle regulatory genes and mediating the cell cycle. The significantly decreased global expression level of miRNAs was discovered in various tumor cells, leading to the disorder of miRNAs function and deprivation of cell cycle inhibition [94]. LncRNAs display cancer-related expression profiles based on tumor-specific features. Specifically, hypoxia is a major cause of cancer progression and chemotherapeutic resistance acquisition, leading to aberrant expression of several lncRNAs. LncRNA p21 is hypoxia-responsive that develops a positive feedback loop with HIF-1α to motivate glycolysis in cancer [95]. Upregulation of the hypoxia-inducing lncRNA EFNA3 accelerates Ephrin-A3 accumulation at the cell surface to promote tumor invasion and metastasis [96]. Widespread dysregulation of circRNAs has been discovered in multiple cancers, which is frequently accompanied by reduced global circRNA levels in rapidly proliferating cancer cells, indicating that many circRNAs act as tumor suppressors. However, individual circRNAs could be upregulated in cancer cells to promote oncogenesis because their slow generation and high stability guarantee their accumulation in non-proliferative cells [97-100].
4 METABOLIC REWIRING AFFECTS EPIGENETICS THROUGH REGULATING SUBSTRATES AND COFACTORS AVAILABILITY OF CHROMATIN REGULATORS
Many metabolites serve as substrates or cofactors for chromatin-modifying enzymes, and their cellular concentration ranges overlap with the kinetic parameters of these enzymes [101]. Therefore, the availability of these critical metabolites could influence the activities of chromatin-modifying enzymes and, thus, the abundance of epigenetic modifications. However, chromatin remodelers are saturated with their substrate, ATP, because of the high intracellular ATP concentration. Their activities are thus generally unaffected by metabolic reprogramming [102]. We think these are general mechanisms explaining how metabolism controls epigenetics in cancer. Researches have revealed that metabolism could regulate tumor initiation, differentiation, proliferation, metastasis, and drug resistance through epigenetics. That is to say, these intricate interactions function in almost all stages of tumorigenesis, even before the malignant transformation. One representative example is that metabolic regulation of the epigenome drives tumorigenesis in posterior fossa A ependymomas. Hypoxia induces metabolic reprogramming, significantly decreasing SAM levels while increasing α-KG and acetyl-CoA levels. The perturbations of these three key metabolites attenuate the substrate availability of H3K27 methyltransferases, promoting the activity of H3K27 demethylases, and fueling H3K27 acetyltransferases. Collectively, these changes lead to a unique epigenetic landscape characterized by H3K27 hypomethylation and hyperacetylation [103]. How the aforementioned key metabolites, along with other primary metabolites, build a bridge between aberrant metabolism and the epigenome in cancer will be discussed in detail below.
We have also gained some new insights into cancer metabolism beyond conventional wisdom. First, cancer metabolism is subcellularly compartmentalized, which allows metabolites to participate in many distinct biological processes [104]. Several metabolic intermediates, such as acetyl-CoA and NAD+, can be produced in the nucleus. Recent research has shown that almost all TCA cycle-associated enzymes exist in the nucleus, forming a local metabolic pool [105]. Thus, the concentration of these metabolites is regulated by, but relatively independent of, mitochondrial and cytoplasmic metabolism. This represents an additional mechanism that tumor cells can exploit to regulate chromatin. Second, newly identified histone post-translational modifications, such as histone lactylation and succinylation, are also metabolically sensitive [106, 107]. They orchestrate two of the most important metabolic pathways, glycolysis and TCA cycle, and epigenetic transcriptional responses. To delve further into these histone modifications will be very interesting.
4.1 Substrates of chromatin modifiers
Acetyl-CoA is a crucial metabolite in many cellular compartments. It is mainly produced by pyruvate oxidative decarboxylation, fatty acid β-oxidation, and branched amino acid catabolism in the mitochondrial matrix [108]. Acetyl-CoA cannot penetrate the mitochondrial membrane directly. Instead, it forms citrate with oxaloacetate in the TCA cycle, which can be transported into the cytosol and decomposed to acetyl-CoA by ATP-citrate lyase (ACL) [109]. Acetate metabolism catalyzed by acetyl-CoA synthetase 2 (ACSS2) is an alternative source of cytosolic acetyl-CoA [108]. Histone acetylation relies on the acetyl-CoA synthesis and can be dynamically regulated by fluctuating concentrations of cellular acetyl-CoA derived from glucose and lipids under physiological conditions [110-113].
Metabolic reprogramming could alter the ratio of acetyl-CoA to coenzyme A and subsequently affect histone acetylation states in cancer cells. AMPK is responsible for promoting glycolysis and the TCA cycle in leukemia. AMPK promotes the production of acetyl-CoA, which maintains global histone acetylation to facilitate the expression of leukemogenic transcription factors [114]. The PI3K/AKT pathway is activated in human prostate cancer and gliomas. AKT activity correlates with histone acetylation levels in clinical samples. KRAS mutations promote acetyl-CoA production and histone acetylation by phosphorylating ACL and enhancing glucose uptake in an AKT-dependent manner [115]. AKT-induced ACL and histone acetylation are also required for acinar-ductal metaplasia and pancreatic tumorigenesis. Reduced acetyl-CoA levels caused by ACL ablation impair early pancreatic tumorigenesis [116]. The ACL is augmented in melanomas. ACL regulates MITF transcription and promotes melanoma growth through P300-mediated histone acetylation. Targeting ACL increases the sensitivity of MAPK inhibition in BRAF-mutant melanoma [117]. ACL is essential for maintaining global histone acetylation, whereas ACSS2 can compensate for acetyl-CoA levels in a dose-dependent manner when ACL is deficient [118]. Acyl-CoA thioesterase 12 (ACOT12) could hydrolyze acetyl-CoA into acetate and coenzyme A. Downregulated ACOT12 increases acetyl-CoA abundance along with histone H3 acetylation levels in hepatocellular carcinoma (HCC), which epigenetically promote EMT and metastasis [119]. Reprogrammed lipid metabolism is involved in controlling cell state transitions. Enhanced fatty acid oxidation (FAO) contributes to acquiring a mesenchymal phenotype in breast cancer cells by producing acetyl-CoA to maintain histone acetylation on the promoters of genes associated with EMT [120].
These acetyl-CoA-producing enzymes are also located in the nucleus, locally regulating histone acetylation. DNA damage signaling promotes nuclear ACL phosphorylation. Phosphorylated ACL produces acetyl-CoA locally and promotes histone acetylation at double-strand break sites, thereby recruiting BRCA1 and favoring homologous recombination repair. These results indicate that acetyl-CoA production by ACL is spatially and temporally controlled [121]. Growth factors or mitochondrial dysfunction augment pyruvate dehydrogenase complex (PDC) translocation from the mitochondria into the nucleus during the S phase. The nuclear PDC generates acetyl-CoA and promotes the acetylation of H3K9 and H3K18, which supports S phase progression [122]. In Pten deficient prostate tumors, PDC has a strong nuclear localization. The nuclear PDC regulates H3K9ac and thus affects the expression of lipid synthesis genes [123]. This is an alternative way to generate acetyl-CoA for histone acetylation in addition to ACL. However, it is astonishing that silencing ACL and PDC affect different sites of acetylation [122, 123].
Under stress conditions, such as nutrient deprivation or hypoxia, acetyl-CoA generated from glucose is markedly reduced. Specific subsets of cancer cells may be addicted to utilizing acetate as an alternative carbon source for maintaining acetyl-CoA production, which is mediated by ACSS [124-126]. Acetate can restore histone acetylation at H3K9, H3K27, and H3K56. Increased histone acetylation at FASN and ACACA promoter regions promotes de novo lipid synthesis [127]. However, the proportion of exogenous acetate-derived acetyl-CoA used for histone acetylation is relatively low compared to the amount flowing into mitochondrial metabolism and lipogenesis [128, 129]. Under metabolic stress, ACSS2 translocates to the nucleus and maintains cell survival and growth by promoting H3 acetylation at the promoter regions of lysosomal biogenesis and autophagy-related genes. The acetate needed for nuclear ACSS2 to produce acetyl-CoA is generated by histone deacetylation [128]. Nuclear ACSS2 maintains histone acetylation by acetate recapturing, which could explain how cancer cells balance the need for acetyl-CoA and the lack of nutrition [128, 129].
SAM is synthesized from methionine and ATP during the methionine cycle, which is essential for one-carbon metabolism [130]. Serine and other amino acids, such as glycine and threonine, are the major one-carbon unit donors of one-carbon metabolism [24, 131]. Serine can also contribute to SAM production by supporting de novo ATP synthesis to offer adenosine beyond providing one-carbon units for remethylating homocysteine [132]. The methylation status is modulated by cellular SAM levels tuned by one-carbon metabolism [133, 134].
Cancer cells are addicted to serine, which contributes to nucleotide synthesis, methylation, and antioxidant activity. Liver kinase B1 (LKB1) mutation synergizes with KRAS activation to potentiate glycolysis and serine metabolism, which favors SAM production. Elevated SAM generation alters the epigenetic landscape of DNA methylation and dynamically supports retrotransposon methylation and transcriptional silencing. However, it seems to have little effect on histone and RNA methylation levels [135]. SHMT2, the gene encoding serine hydroxymethyltransferase 2 (SHMT2) in serine catabolism, is frequently amplified in B-cell lymphomas. SHMT2 is responsible for converting serine into glycine and contributes a one-carbon unit to the folate cycle. Overexpressed SHMT2 changes the DNA methylation state globally and epigenetically silences tumor suppressor genes in lymphoma [136]. Phosphoglycerate dehydrogenase (PHGDH), the critical enzyme in the de novo serine synthesis pathway, directs glycolytic flux into the one-carbon metabolic network. Upregulated PHGDH increases metabolite levels in the methionine cycle and promotes histone methylation [137]. Small cell/neuroendocrine prostate cancer (NEPC), which is highly aggressive, has a distinct DNA methylation profile from that of adenocarcinoma during differentiation. Protein kinase C λ/ι (PKCλ/ι) deficiency increases the one-carbon metabolism through the mTORC1/ATF4/PHGDH axis to fuel DNA methylation, which promotes NEPC differentiation [138].
Methionine metabolism can also alter SAM and SAH concentrations, thus quantifying histone methylation. Methionine restriction leads to decreased H3K4me3 at promoters and the expression of colorectal cancer-associated genes [134]. Cancer stem cells depend highly on methionine because of their high SAM consumption rate. Inhibition of the key enzyme, methionine adenosyltransferase 2A (MAT2A), in the methionine cycle ablates histone methylation in cancer stem cells, which impairs their tumor formation ability and resistance to cisplatin [139].
Deregulation of nicotinamide N-methyltransferase (NNMT) could alter the epigenetic state by consuming methyl units into 1-methylnicotinamide (1MNA), which consequently attenuates the SAM/SAH ratio. Deregulated NNMT is found in many different tumors and supports tumorigenesis by selectively reducing the histone methylation of several specific genes and increasing their expression [140].
Other metabolites can also act as substrates for histone modifications [141]. Evidence of the role of these histone modifications in cancer continues to emerge. Lactate is a product of the Warburg effect and is a key metabolite and signaling molecule. It plays essential roles in multiple biological processes during tumor progression, such as angiogenesis, immune escape, and cell proliferation [142]. However, their role in chromatin modification has long been overlooked. Recently, researchers have found that histone lactylation derives from lactate and could contribute to gene expression [143, 144]. Active glycolysis provides sufficient lactate for lactylation in ocular melanomas. H3K18la is enriched in YTH N6-methyladenosine RNA binding protein 2 (YTHDF2) promoter regions and promotes the transcription of YTHDF2. As an N6-methyladenosine (m6A) reader, YTHDF2 binds to the m6A sites of PER1 and TP53 mRNAs for degradation [145]. Lactylation provides new insight into the Warburg effect and requires further investigation [146].
4.2 Cofactors of chromatin modifiers
α-KG is an intermediate in the TCA cycle and is produced from isocitrate by isocitrate dehydrogenase (IDH). α-KG is the co-substrate for a class of dioxygenase enzymes such as JHDMs, TETs, and prolyl hydroxylase [147]. In human pluripotent stem cells, α-KG induces histone and DNA demethylation and promotes differentiation [148]. It can be presumed that α-KG has an important role in regulating epigenomic plasticity. The same mechanism could explain the antitumor effects of α-KG. In PDA, p53 inactivation leads to reduced α-KG levels by rewiring the glucose and glutamine metabolism, which impairs TETs activity. This causes tumor cells to gain the characteristics of poor differentiation and high aggressiveness [149]. When exogenous serine is abundant, squamous cell carcinoma (SCC) cells show enhanced mitochondrial pyruvate metabolism and prevent NAD+ regeneration by reducing pyruvate to lactate. Limited NAD+ is not conducive to serine synthesis. Thus, SCC cells inhibit the de novo serine synthesis pathway, resulting in the accumulation of the byproduct, α-KG. Decreased α-KG inhibits histone demethylases and H3K27me3 demethylation, which blocks cancer stem cells from differentiating. This feature maintains the stemness of tumor stem cells and promotes tumor initiation [147].
Glutamine replenishes the TCA cycle to produce α-KG [150]. Increased consumption of glutamine leads to local glutamine deficiency in tumor core regions. Hypermethylation of histones caused by decreased glutamine and α-KG levels causes cancer cell dedifferentiation and BRAF inhibition resistance [151]. Glutamine supplementation increases the downstream metabolite, α-KG. An increase in α-KG concentration could suppress the oncogenic pathway in melanoma by decreasing global H3K4me3 levels and affecting H3K4me3-dependent transcription [152]. However, the role of glutamine in cancer remains unclear. KRAS-mutant colorectal cancer cells show increased reliance on glutamine. Mutant KRAS promotes glutaminolysis and succinate, fumarate, and malate accumulation in the TCA cycle, whereas the level of α-KG decreases. Downregulation of α-KG to succinate ratio inhibits the activities of demethylases and impacts genome-wide DNA and histone methylation. Aberrant methylation patterns activate WNT/β-catenin signaling and increase tumor stemness [153].
NAD+ is a co-enzyme that mediates oxidation-reduction (redox) reactions in many metabolic pathways, including glycolysis, TCA cycle, OXPHOS, and FAO. NAD+ regulates cell metabolism, redox homeostasis, genome stability, and histone modifications [154]. SIRTs remove acyl groups from lysine residues and transfer NAD+ into 2′-O-acyl-ADP ribose (OAADPR) and nicotinamide (NAM) [155]. SIRTs can sense NAD+ levels, and their activity may be modulated by cellular concentrations of NAD+ and NAM [156, 157].
The metabolic switch from FAO to glycolysis decreases NAD+ concentration and inhibits SIRT1, thereby blocking H4K16 deacetylation in skeletal muscle stem cells. This directly shows that metabolic reprogramming can rewrite the epigenetic state through NAD+ [156]. For breast cancer cells, nicotinamide phosphoribosyltransferase (NAMPT) and NMN adenylyltransferase 1 (NMNAT1) regulate specific gene expression in a SIRT1-dependent way. As the key enzymes of the NAD+ salvage pathway, NAMPT and NMNAT1 regulate NAD+ concentration and SIRT1 deacetylation activity, thus affecting H4K16ac levels at gene promoters. SIRT1 can recruit NMNAT1 to target gene promoter regions, creating a locally high NAD+ concentration to control SIRT1 activity [158]. In melanoma, the BRAF/ERK/STAT5 pathway transcriptionally regulates NAMPT expression. Overexpressed NAMPT changes the histone modification landscape and allows melanoma cells to switch to a more invasive phenotype associated with resistance to targeted therapies and immunotherapies [159].
4.3 Oncometabolites: competitive inhibitors of chromatin modifiers
In cancer cells containing mutated metabolic enzymes, 2-hydroxyglutarate (2-HG), fumarate, and succinate may be produced and accumulate [160]. It is worth noting that 2-HG is chiral and exists as the two isoforms, D2-HG and L2-HG. These two enantiomers are differentially upregulated in distinct tumor contexts. These abnormal metabolites mix into the metabolic pool and competitively inhibit the activity of α-KG-dependent dioxygenases, such as multiple histone demethylases and the TET family of 5-methylcytosine hydroxylases, because of their similar structure to α-KG [161, 162]. They are also called oncometabolites because their aberrant accumulation can promote malignant transformation [160]. For example, IDH1/2 encodes isocitrate dehydrogenase 1/2, which usually catalyzes the oxidative decarboxylation of isocitrate to α-KG. Mutated IDH1/2 gains the function of producing 2-HG, specifically the D enantiomer, from α-KG [163, 164]. Emerging evidence indicates that elevated 2-HG levels could alter global histone and DNA methylation patterns and drive tumorigenesis in leukemia and glioma [165-167].
Impaired histone and DNA demethylation are associated with blocked cell differentiation [16, 168-173]. For example, IDH2 mutation impairs the differentiation potential of multipotent cells and endows them with the ability to escape contact inhibition. IDH mutations are sufficient to promote malignant transformation and generate poorly differentiated sarcomas [174]. IDH mutations also cause genome-wide DNA hypermethylation at the cohesin- and CTCF-binding sites. Decreased CTCF binding widely compromises chromosomal topology and results in oncogenes like PDGFRA aberrant activation through interaction with distant enhancers [175]. IDH mutations alter cell metabolism and DNA repair through epigenetic mechanisms. Mutant IDH silences lactate dehydrogenase A (LDHA) by increasing promoter methylation [176]. D2-HG increases repressive histone methylation marks at the ATM promoter, resulting in impaired DNA damage repair and self-renewal of hematopoietic stem cells (HSCs) [177]. There are some similar findings in gliomas and acute myeloid leukemia (AML) that IDH1/2 mutations induce homologous recombination defects and sensitize tumor cells to poly (ADP-ribose) polymerase inhibition [178]. Besides, mutant IDH produces D2-HG and epigenetically suppresses the expression of interferon γ response genes, which impedes immune response in cholangiocarcinoma [179].
Under physiological conditions, the L enantiomer of 2-HG is produced by LDHA and malate dehydrogenase 1 and 2 (MDH1/2) in response to hypoxia stress [180-182]. It has a far more potent inhibitory effect on α-KG-dependent dioxygenases than the D enantiomer [161, 162]. L2-HG, rather than D2-HG, mainly elevates in renal cell carcinoma (RCC) due to reduced expression of L2-HG dehydrogenase (L2HGDH), which can convert L2-HG back into α-KG to avoid the accumulation of L2-HG. Consistently, accumulation of L2-HG reduces DNA 5hmC and increases repressive trimethylated histone marks like H3K9me3 and H3K27me3 [183]. Restoring L2HGDH can stunt tumor growth [184].
In addition to IDH, mutations in succinate dehydrogenase (SDH) and fumarate hydratase (FH) have been identified. They may share the same oncogenic mechanism. FH and SDH mutants lose their enzymatic activities and lead to fumarate and succinate accumulation, inhibiting α-KG-dependent dioxygenases [185]. SDH-mutant gastrointestinal stromal tumors (GIST), paragangliomas, and FH-mutant renal cell carcinomas show characteristic hypermethylation patterns [186-190]. In paraganglioma, hypermethylated and downregulated genes are involved in chromaffin cell differentiation and EMT [187]. Consistent with findings in IDH-mutant gliomas, abnormal DNA methylation at CTCF sites in SDH-deficient GIST compromises FGF and KIT insulators, reorganizes chromosome topology, and allows super-enhancers to interact with and activate oncogenes [191]. Fumarate and succinate accumulation suppresses homologous recombination DNA repair by inhibiting KDM4A and KDM4B and makes tumor cells vulnerable to PARP inhibitors [192, 193].
When it comes to the mutation of enzymes in the TCA cycle, another essential and ubiquitous post-translational modification, succinylation, is also affected. Succinyl-CoA, the substrate of succinylation reaction, is mainly generated from the TCA cycle. Histone succinylation can be mediated both enzymatically and non-enzymatically. KAT1 and KAT2A are responsible for depositing histone succinylation marks, whereas SIRT5 and SIRT7 are histone desuccinylases [194-196]. Histone succinylation is generally associated with transcriptional activation and broadly regulates the expression of tumor-related genes [197-200]. KAT2A interacts with the α-ketoglutarate dehydrogenase (α-KGDH) complex in the nucleus. α-KGDH complex locally catalyzes succinyl-CoA production and fuels KAT2A-mediated H3K79 succinylation, which induces gene expression and promotes tumor growth [197]. In IDH1/2-mutated gliomas, inhibition of SDH and subsequent accumulation of succinyl-CoA are caused by D2-HG, which foster widespread histone and nonhistone protein hypersuccinylation in different cellular compartments. Although hypersuccinylation induced by oncometabolites preferentially impacts mitochondrial metabolism, it also profoundly affects chromatin [201]. SDH loss selectively perturbs genome-wide chromatin succinylation in promoter regions. Genes involved in transcriptional regulation and RNA processing are most affected [202]. However, many tumors, including esophageal squamous cell carcinoma (ESCC), are globally hyposuccinylated. It suggests that the functions of histone succinylation are context-dependent [203]. Limited researches have provided a glimpse into how succinyl-CoA is used explicitly by the tumor to alter the epigenetic chromatin state. Further detailed studies are urgently needed to unravel this important link (Figure 2).
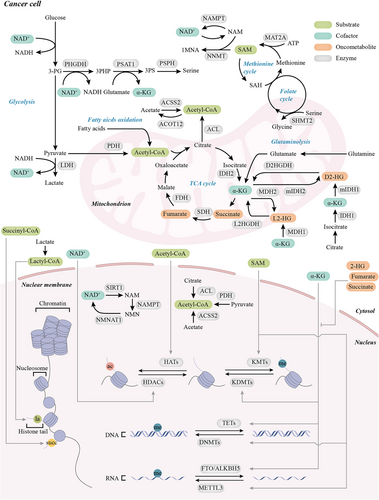
Aberrant epigenetic modifications have previously been attributed to mutation and abnormal expression of epigenetic enzymes. Cellular metabolism, which provides substrates, cofactors, and oncometabolites for epigenetic enzymes, also dynamically affects the epigenetic landscape. This fundamental process is precisely controlled under normal circumstances. However, these “molecular signals” can be excessive, insufficient, and even erroneous in cancer. Merely inhibiting a specific metabolic pathway or epigenetic enzyme will activate compensating mechanisms. It is conceivable that resistance to monotherapies is almost inevitable. The results presented above provide the molecular bases for the necessity of targeting the intersections between metabolism and epigenetics in cancer. Simultaneously targeting both upstream and downstream epigenetic enzymes of the metabolic-epigenetic axis may achieve much more significant and durable responses.
In addition to being confirmed in preclinical studies, this concept has exhibited promising clinical results in treating leukemia. IDH-mutant leukemia possesses a hypermethylated phenotype. Although hypomethylating agents and IDH inhibitors have been approved by authorities and improved the clinical outcomes of AML patients, drug resistance invariably occurs. Blocking the source of 2-HG (IDH inhibitor, ivosidenib) coordinates synergistically with the inhibition of DNA methyltransferase (DNMT inhibitor, azacytidine) in patients unable to receive intensive induction chemotherapy. Combined therapy significantly improved drug responses, event-free survival, and overall survival compared to azacytidine monotherapy. Toxic effects were durable. These important findings may eventually offer a new treatment option to AML patients with IDH mutations [204, 205].
5 ABERRANT EPIGENETIC PATTERNS CONTRIBUTE TO METABOLIC REPROGRAMMING
Genetic and epigenetic alterations actively participate in the metabolic reprogramming of cancer. For example, oncogenic Kras mutations selectively rewire glucose metabolism to promote pancreatic tumor growth [3]. Compared with genetic mutations, epigenetic regulations are reversible and variable. Epigenetic modifiers modulate metabolism by directly changing the transcriptional activities of metabolic enzymes or proteins in metabolism-related signaling pathways according to the needs of tumor cells. Increased histone and DNA methylation mark transcriptionally repress fructose-1,6-biphosphatase (FBP1), which triggers the reprogramming of glucose metabolism to sustain cancer stem cell-like properties in breast cancer cells [206]. The roles of ncRNAs in regulating metabolic reprogramming are much more complicated, involving both transcriptional and post-transcriptional regulations. Exploring the epigenetic roles of ncRNAs in regulating metabolism will dramatically expand the list of drug targets. Although studies are emerging, there remain important unanswered questions. One outstanding issue is how these epigenetic processes are coordinated to promote tumor development by regulating metabolism.
Here, we introduce the four pivotal epigenetic mechanisms and discuss their contributions. Given that many recurrent mutations in epigenetic regulators have been identified as cancer driver mutations, their roles in promoting cancer metabolism will be highlighted.
5.1 DNA modifiers and modification
Abnormal methylation of promoter DNA occurs in metabolic enzymes. The TET3 protein is often upregulated in AML cells. TET3 induces the expression of glucose metabolism-related genes by depositing 5hmC marks on their promoters [207]. Hypomethylation of the promoter contributes to the upregulation of hexokinase 2 (HK2) in liver cancer and glioblastoma. Enhanced HK2 levels promote increased glycolytic flux [208, 209]. DNMT1 downregulates FBP1 in basal-like breast cancer by binding and methylating the FBP1 promoter, inhibiting gluconeogenesis and enhancing cancer cell glycolytic rates [206]. The glucose transporter (GLUT) plays an essential role in glucose metabolism in cancer. Elevated GLUT promotes glucose access to tumor cells and facilitates aerobic glycolysis. Consequently, lactate and pyruvate, metabolites of aerobic glycolysis, acidify the tumor microenvironment and increase tumor proliferation and invasion. Promoter hypermethylation causes the inactivation of DERL3, a crucial regulator of the endoplasmic reticulum-associated protein degradation pathway, which enhances the expression of GLUT1 and promotes aerobic glycolysis. This is mediated by DNMT1 and DNMT3B [210]. In addition, elevated CAV-1 expression by hypomethylation of the promoter CpG site upregulates GLUT3 transcription, stimulates glucose uptake, and increases aerobic glycolysis [211].
5.2 Histone modifiers and modifications
Loss of histone methyltransferase EZH2 synergizes with oncogenic NRAS mutations to accelerate leukemic transformation in myeloid neoplasms. In terms of mechanism, EZH2 epigenetically silences branched-chain amino acid transaminase 1 (BCAT1) and disturbs branched-chain amino acids (BCAAs) metabolism in hematopoietic stem/progenitor cells (HPSCs). Loss of EZH2 abolishes promoter repression and activates enhancers of BCAT1, leading to the accumulation of BCAAs and the subsequent activation of mTOR signaling in leukemia-initiating cells [212]. The histone methyltransferase KMT2D is frequently mutated in lung cancer. KMT2D deficiency promotes lung tumorigenesis and upregulates glycolysis by impairing super-enhancers of PER2 [213]. In melanoma, KMT2D loss causes genome-wide reduction of H3K4me1-marked active enhancer chromatin states and subsequently activates IGF1R/AKT to increase glycolysis [214]. KMT2D is transcriptionally repressed and mutated in pancreatic cancer. KMT2D repression promotes a metabolic shift to glycolysis and alters the cellular lipid profile of pancreatic cancer cells, which provides energy for cell proliferation [18]. Overexpressed histone methyltransferase NSD2 establishes H3K36me2 marks at the promoters of genes associated with glucose metabolism to upregulate the expression of HK2, glucose-6-phosphate dehydrogenase (G6PD), and TIGAR in breast cancer. As a result, glucose flux through PPP and NADPH production is upregulated to alleviate reactive oxygen species (ROS) and promote drug resistance [215]. Mutation and activation of histone methyltransferase SETD2 are frequently observed in renal cancer. SETD2-deficient cancer cells exhibit enhanced OXPHOS and fatty acid synthesis [216]. The histone H3K9 methyltransferase G9A (KMT1C) is elevated in many types of cancer and promotes tumorigenesis. G9A activates the serine-glycine biosynthetic pathway by transcriptionally upregulating key enzymes, such as PHGDH, phosphoserine aminotransferase 1 (PSAT1), SHMT2, and phosphoserine phosphatase (PSPH), by increasing H3K9me1 levels around the transcriptional start sites [217]. Consistently, KDM4C, the histone demethylase responsible for removing the repressive mark H3K9me3, could epigenetically coordinate the regulation of amino acid metabolism with G9A. Decreased H3K9me3 level with a concomitant increased ratio of H3K9me1 to H3K9me3 at the promoters of genes associated with the synthesis and transport of seine and glycine promote tumor proliferation [218]. LSD1 (KDM1A) activates glycolysis and represses mitochondrial metabolism and FAO in hepatocellular cancer. H3K4 demethylation in the promoter regions of PGC-1α and LCAD partially explains the mechanism underlying this metabolic preference [219]. KDM5A specifically removes the active mark H3K4me3 on MPC-1 genes in PDA. MPC-1 promotes pyruvate metabolism in mitochondria. Transcriptional inhibition of MPC-1 endows PDA with reliance on glycolysis [220].
P300/CBP regulates the alteration of cancer metabolism and the transcription of enzymes in glycolysis-related metabolic pathways, such as amino acid metabolism, fatty acid metabolism, and nucleotide synthesis, by acetylating histone H3K18/K27 directly at the promoters of metabolic genes [221]. SIRT6 is deleted or downregulated in many cancer types, such as pancreatic and colorectal cancer. The deficiency of SIRT6, the co-repressor of HIF-1α and MYC, promotes tumorigenesis by supporting glycolytic switch, ribosome biogenesis, and glutamine metabolism without activating other oncogenic signaling pathways. Inhibition of glycolysis in SIRT6-deficient cells completely inhibits tumor formation [222, 223]. Mechanistically, SIRT6 deletion, transcriptional silencing, and point mutations cannot deacetylate H3K9 and H3K56 and repress glycolytic gene expression [223, 224]. HDAC11 removes H3K9ac on the LKB1 promoter and inhibits its expression. LKB1 inhibition promotes glycolysis and maintains the stemness of HCC cells [225].
5.3 Chromatin remodeling complexes
Several studies have suggested that the SWI/SNF complex is involved in the rewiring of cancer metabolism. ARID1A, along with other core subunits, can directly bind to the promoter of GLS1. ARID1A inactivation increases the accessibility of the GLS1 promoter and upregulates glutaminase (GLS) expression. ARID1A-inactivated clear cell ovarian carcinoma cells show dependence on glutamine metabolism for aspartate generation, nucleotide synthesis, and a decrease in glucose consumption [226]. Another study found that ARID1A deficiency in ovarian cancer cells impairs the recruitment of SWI/SNF to the transcription start site of SLC7A11 and subsequently reduces cystine uptake and reduced glutathione (GSH) synthesis. Inhibiting the glutamate-cysteine ligase synthetase catalytic subunit (GCLC), a rate-limiting enzyme in the GSH metabolic pathway, induces oxidative stress and the death of cancer cells. Nevertheless, ARID1A-deficient ovarian cancer cells are insensitive to GLS1 inhibition [227]. SMARCA4 is frequently mutated and inactivated in lung adenocarcinoma. SMARCA4 regulates genes in the hypoxic response pathway and glycolysis to combat energy stress. However, augmented fatty acid and protein synthesis in SMARCA4-mutant cells results in substantial energy demand. Inconsistent with the Warburg effect, defective glycolytic capacity drives SWI/SNF-mutant lung adenocarcinoma tumors to shift energy metabolism from glycolysis to OXPHOS [19]. Elevated BRG1 (SMARCA4) increases fatty acid synthesis in breast cancer by transcriptionally activating lipogenic genes, such as ACC, FASN, ACL, and ACSL1. Upregulated de novo lipogenesis can greatly promote tumor proliferation [228].
The above studies summarize the link ATP-dependent CRCs to cancer metabolism and demonstrate a novel mechanism of how mutant CRCs components contribute to tumorigenesis. Remarkably, these findings provide a new perspective that the vulnerability of SWI/SNF-mutant tumors to metabolism could be a therapeutic target (Table 1).
| Epigenetic regulator | Cancer type | Metabolic alteration |
|---|---|---|
| DNA modifier | ||
| TET3 overexpression [207] | Leukemia | Upregulating glucose metabolism |
| Histone modifier | ||
| EZH2 deficiency [212] | Leukemia | Activating branched-chain amino acids metabolism |
| KMT2D deficiency [213] | Lung cancer | Upregulating glycolysis |
| KMT2D deficiency [214] | Melanoma | Upregulating glycolysis |
| KMT2D inhibition [18] | Pancreatic cancer | Upregulating glycolysis and lipids metabolism |
| NSD2 overexpression [215] | Breast cancer | Upregulating pentose phosphate pathway |
| SETD2 deficiency [216] | Renal cancer | Upregulating oxidative phosphorylation and fatty acid synthesis |
| G9A overexpression [217] | Osteosarcoma, Neuroblastoma, etc. | Upregulating serine-glycine biosynthetic pathway |
| LSD1 overexpression [219] | Liver cancer | Upregulating glycolysis |
| KDM4C overexpression [218] | Cervical cancer, Neuroblastoma, etc. | Upregulating amino acids metabolism |
| KDM5A overexpression [220] | Pancreatic cancer | Upregulating glycolysis |
| P300/CBP overexpression [221] | Liver cancer | Upregulating glycolysis and amino acids metabolism |
| SIRT6 deficiency [222] | Pancreatic cancer, Colorectal cancer, etc. | Upregulating glycolysis |
| HDAC11 overexpression [225] | Liver cancer | Upregulating glycolysis |
| Chromatin remodeler | ||
| ARID1A deficiency [226] | Ovarian cancer | Upregulating glutamine metabolism |
| ARID1A deficiency [227] | Ovarian cancer | Inhibiting reduced glutathione synthesis |
| SMARCA4 deficiency [19] | Lung cancer | Upregulating oxidative phosphorylation |
| SMARCA4 overexpression [228] | Breast cancer | Upregulating fatty acids synthesis |
- Abbreviations: TET3, Ten-eleven translocation family protein 3; EZH2, Enhancer of zeste homolog 2; KMT2D, Histone lysine methyltransferase 2D; NSD2, Nuclear receptor binding SET domain protein 2; SETD2, SET domain containing 2; G9A, Euchromatic histone lysine methyltransferase 2; LSD1, Lysine-specific demethylase 1; KDM4C, Histone lysine demethylase 4C; KDM5A, Histone lysine demethylase 5A; P300/CBP, E1A binding protein p300/CREB binding protein; SIRT6, Sirtuin 6; HDAC11, Histone deacetylase 11; ARID1A, AT-rich interacting domain-containing protein 1A; SMARCA4, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4.
5.4 Non-coding RNAs
MicroRNAs regulate gene expression at the post-transcriptional level [229]. The role of miRNAs in metabolism has been thoroughly investigated and documented; consequently, it is not discussed in detail in this section [230, 231]. Here, we emphasize that miRNAs are indispensable coordinators of metabolic regulatory networks.
Long non-coding RNAs (lncRNAs) participate in various physiological and pathological processes. LncRNAs are involved in various important cellular processes and play pivotal roles in gene regulation at multiple levels [232].
LncRNAs are involved in cancer metabolism via diverse mechanisms. LncRNAs can recruit chromatin modifiers to target genes and alter their epigenetic status. LINC00184 recruits DNMT1 to the PTEN promoter, increasing the methylation level of the PTEN promoter and inhibiting the expression of PTEN [233]. Fusobacterium nucleatum, an oncobacterium, activates glycolysis in colorectal cancer by increasing lncRNA ENO1-IT1. LncRNA ENO1-IT1 interacts with KAT7 specifically and mediates KAT7 binding to the promoter region of ENO1. Increased H3K27Ac levels promote transcription of enolase 1 (ENO1), which increases tumor glucose metabolism and progression [234]. LncRNAs can regulate gene expression by interfering with transcription. In prostate cancer, lncRNA PCGEM1 occupies DNA loci on the promoters of metabolic genes involved in glucose, lipid, and glutamine metabolism that overlap with c-Myc. LncRNA PCGEM1 promotes the recruitment of c-Myc to its target genes and induces transactivation activities. These results emphasize that the lncRNA PCGEM1 is a vital transcriptional regulator in restructuring metabolic networks [235]. LncRNAs also bind to other transcription factors, AHR, GLI2, and E2F1, to promote metabolic switching, thereby stimulating tumor progression [236-238].
LncRNAs mediate the splicing, degradation, and translation of mRNA. The lncRNA CCAT2 alters metabolism by facilitating glycolysis and glutaminolysis. The lncRNA CCAT2 acts as a scaffold binding GLS pre-mRNA and CFIm complex and regulates alternative splicing of GLS in an allele-specific manner. Moreover, other metabolic pathways, such as carbohydrate metabolism and fructose and mannose metabolism, may share the same alternative splicing mechanism [239]. LncRNA LNCAROD interacts with SRSF3, a splicer that mediates alternative splicing of PKM. Splicing switching of PKM from PKM1 to PKM2 upregulates glycolysis in HCC [240]. LncRNA GLS-AS, an intronic antisense lncRNA, is derived from GLS. It can form double-stranded RNA with GLS pre-mRNA and recruit the ADAR/Dicer complex, which silences GLS expression. Under nutritional stress conditions, downregulated lncRNA GLS-AS causes pancreatic cancer to accommodate glutamine and glucose deprivation [241]. Trastuzumab-resistant breast cancer cells have upregulated lncRNA AGAP2-AS1. LncRNA AGAP2-AS1 forms a complex with HuR, which binds to and stabilizes carnitine palmitoyl transferase 1 (CPT1) mRNA to improve its expression, promote FAO, and induce drug resistance [242]. LncRNAs can mediate c-Myc mRNA decay and glycolysis by virtue of IGF2BPs [243-245].
LncRNAs can regulate gene expression as sponges of miRNAs. LncRNA PVT1 contains miRNA-complementary sites and acts as a competing endogenous RNA (ceRNA) of miR-143, which targets and suppresses HK2 in gallbladder cancer. The sequestration of miR-143 by lncRNA PVT1 elevates HK2 expression and facilitates the Warburg effect and gallbladder cancer progression [246]. This is the most extensively studied mechanism of the lncRNA-mediated metabolic switch. The same mechanism fundamentally applies to aberrant regulation of metabolic transporters, key enzymes, and transcription factors associated with glucose, glutamine, and fatty acid metabolism [240, 242, 247-250]. LncRNAs can bind to metabolic enzymes or transcriptional factors and modulate their activity or block their post-translational modifications. LncRNA HULC repositions PKM2 and LDHA to the cell membrane and enhances the interaction between these glycolytic enzymes and their phosphorylation regulator, FGFR1. FGFR1 modulates enzymatic activities and promotes glycolysis by elevating their phosphorylation levels [251]. Hypoxia-induced lincRNA-p21 competitively binds to VHL and prevents hydroxylated HIF-1α from interacting with it. Disassociation from VHL prevents HIF-1α from degradation via the VHL-dependent ubiquitin-proteasome pathway [95]. In triple-negative breast cancer, LINK-A recruits BRK to phosphorylate HIF-1α at Tyr565. Phosphorylation of Tyr565 attenuates the Pro564 site hydroxylated by PHD1 [252]. Many other lncRNAs stabilize PKM2, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3), and c-Myc by directly binding and blocking these proteins from ubiquitination-mediated degradation [253-256]. LncRNAs are found to function as scaffolds for proteins and RNA to form condensates. Under glutamine deprivation, lncRNA GIRGL forms a complex with CAPRIN1 and GLS1 mRNA and promotes the formation of stress granules via liquid-liquid phase separation. This process contributes to the translational suppression of GLS, which favors tumor growth in a glutamine-restricted environment [257].
Circular RNAs (circRNAs) have a single-stranded, covalently closed-loop structure. Growing evidence indicates that circRNAs play crucial roles in many diseases and have multiple biological functions [258]. Mechanistically, circRNAs can function as ceRNAs to sponge miRNAs and regulate downstream targets. Additionally, circRNAs can regulate transcription, interact with proteins, or even be translated into peptides [87].
Some circRNAs have been identified as key participants in reprogramming cancer metabolism. The overwhelming majority of research has focused on their ability to act as molecular sponges, which could antagonize the regulation of metabolic enzymes, transcription factors, and signaling pathways by miRNA. In HCC, miR-338-3p represses glycolysis by targeting and degrading PKM2. CircMAT2B sponges miR-388-3p and promotes glucose metabolism reprogramming and tumor cells’ malignancy under hypoxic conditions [259]. CircENO1 upregulates ENO1 and modulates glycolysis by targeting miR-22-3p in lung adenocarcinoma (LUAD) [260]. In pancreatic cancer, circMBOAT2 favors glutaminolysis by sponging miR-433-3p and upregulating glutamic-oxaloacetic transaminase 1 (GOT1)[261]. Upstream molecules modulate glycolysis like HIF-1α [262, 263], PTK [264], and c-Myc [265], and upstream molecules related to glutamine metabolisms, such as Wnt2 [266], USP5 [267], and IGF [268], are also found to be regulated by the circRNA-miRNA axis.
CircRNAs can directly bind to target mRNA and regulate gene expression at the transcriptional level. CircRNF13 is a tumor suppressor that targets and stabilizes SUMO2 mRNA. SUMO2 accelerates GLUT1 degradation by promoting its SUMOylation and ubiquitination. Downregulated circRNF13 enhances aerobic glycolysis in nasopharyngeal carcinoma (NPC) [269].
Various modes of circRNA-protein interactions are newly clarified mechanisms responsible for metabolic rewiring, which have not been thoroughly studied [270]. CircACC1 is induced under metabolic stress and plays a critical role in AMPK-mediated metabolic reprogramming in colorectal cancer. CircACC1 binds to the β1 and γ1 subunits of AMPK and facilitates holoenzyme assembly and stability. AMPK phosphorylates and inactivates ACC1 to increase fatty acid β-oxidation but has the opposite effect on 6-phosphofructo-2-kinase (PFK2) to promote glycolysis [271]. CircCUX1 binds to EWSR1 and promotes its interaction with MAZ. Activated MAZ promotes the transcription of CUX1, a transcription factor that facilitates glycolysis [272]. CircCDKN2B-AS1 recruits IMP3 (IGF2BP3) to the HK2 mRNA, making it more stable. Increased expression of HK2 favors glycolysis in cervical cancer [273]. In colorectal cancer, circMYH9 impedes the binding between hnRNPA2B1 and p53 pre-mRNA. CircMYH9 relieves transcriptional repression of serine and glycine anabolism by impairing the expression of p53 [274]. In LUAD, circDCUN1D4 forms a ternary complex with HUR and TXNIP mRNA and regulates glycolysis in a TXNIP-dependent manner [275] (Figure 3).
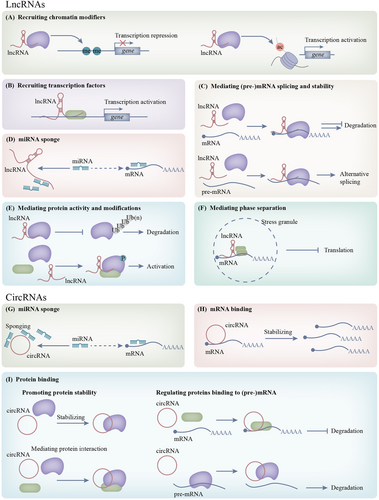
Epigenetic modifications, chromatin remodeling, and ncRNAs participate in the precise regulation of metabolism to favor tumor initiation and progression. They control the ability of tumor cells to uptake nutrients, metabolize nutrients, and adapt to nutrition deprivation. Dysregulated epigenetic patterns can cause specific metabolic preferences or dependencies in tumor cells. These weaknesses can be exploited and directly targeted. Furthermore, epigenetic drugs may profoundly remodel cellular metabolic states and thus sensitize tumor cells to other metabolic drugs. One such example is that dual inhibition of DNMT and KMT reverses the Warburg effect and causes OXPHOS dependence in glycolysis-addicted hematological malignancies [276]. Targeting mitochondrial metabolic stress potentiates the effects of epigenetic drugs. This drug combination shows encouraging results in the clinical trial. In older patients with AML, azacitidine plus venetoclax, a BCL2 inhibitor, significantly improved the median overall survival to 14.7 months, as compared with 9.6 months in the group with azacitidine alone [277]. These basic and clinical studies may open new avenues for developing combination strategies based on epigenetic and metabolic drugs.
6 EMERGING ROLES OF RNA EPIGENETICS IN CANCER METABOLISM
Dynamic RNA modification is an emerging research field termed “RNA epigenetics” [278, 279]. Prevalent modifications on mRNA include m6A, N7-methylguanosine (m7G), 5-methylcytosine (m5C), N1-methyladenosine (m1A), pseudouridine (Ψ), inosine (I), and uridine (U). m6A is the most abundant epigenetic mRNA modification, accounting for 60% of RNA methylation. M6A RNA modifications regulate mRNA splicing, nuclear transport, translation, and degradation [280]. As a reversible chemical modification, m6A could also be deposited by writer proteins, removed by eraser proteins, and recognized by reader proteins [281]. M6A is found to regulate gene expression in various biological processes, and disturbed distribution or abundance of m6A could even drive many diseases [282-284]. Accumulating evidence has demonstrated that m6A RNA modification is affected by cancer metabolism; conversely, it extensively impacts cancer metabolic rewiring by modulating the expression of metabolic genes, which drive tumor development. Although there is a lack of relevant studies in the literature, we could envisage that other novel RNA modifications, such as m5C, m1A, and Ψ, are also closely linked with metabolism in cancer. Elucidating the roles of the crosstalk between RNA epigenetics and cancer metabolism will be an important area for further investigation.
In addition to DNA and histone methylation, SAM is also required for RNA methylation. mTORC1 promotes methionine metabolism and increases SAM production via MAT2A, a crucial target for activated mTORC1 signaling. Nevertheless, mTORC1-dependent regulation of SAM synthesis has little impact on DNA and histone methylation states. Tumors with hyperactivated mTORC1 depend on MAT2A-mediated m6A RNA for protein synthesis and cell proliferation [285]. Methylenetetrahydrofolate dehydrogenase 2 (MTHFD2), a mitochondrial enzyme involved in one-carbon metabolism, is elevated in clear cell renal cell carcinoma (ccRCC). MTHFD2 depletion results in decreased global methylation levels of nucleic acids and histones, of which RNA methylation is the most influenced. Increased methylation of HIF-2α mRNA enhances its translation and subsequently promotes aerobic glycolysis [286]. Similar to DNA and histone disturbances, RNA methylation is significantly elevated in IDH-mutant tumors because fat mass and obesity-associated protein (FTO) are α-KG-dependent dioxygenases that can also be competitively inhibited by R-2HG (D2-HG) [287, 288]. However, R-2HG-induced hypermethylation produces contradictory effects on tumorigenesis. In IDH-mutant leukemia, the decreased m6A demethylase activity of FTO abrogates m6A /YTHDF2-mediated upregulation of PFKP and LDHB, attenuates aerobic glycolysis, and inhibits leukemogenesis [289] (Figure 1).
High methyltransferase-like 3 (METTL3) expression increased HK2 and GLUT1 expression depending on its m6A methyltransferase activity. M6A modification regulates HK2 and GLUT1 mRNA levels and stability and is closely correlated with the activation of glycolysis in colorectal cancer [290]. In cervical and liver cancer cells, m6A positively regulates glycolysis by stabilizing and promoting the translation of PDK4, which controls glucose flux into glycolysis and OXPHOS [291]. Another potential target is ENO1 in LUAD [292]. FTO has a synthetic lethal interaction with VHL tumor suppressor in ccRCC. VHL-deficient tumor cells are addicted to glutamine. Increased FTO rewires the metabolic reprogramming and survival of VHL-deficient ccRCC cells by diminishing m6A methylation and enhancing the expression of the glutamine transporter SLC1A5 [293].
Some key transcription factors or upstream regulators related to metabolic reprogramming are also affected by m6A RNA modifications. METTL3 activates glycolysis by promoting m6A modification of HDGF mRNA in gastric cancer [294], HIF-1α mRNA in liver cancer [295], APC mRNA in ESCC [296], and USP48 mRNA in liver cancer [297]. METTL3 enhances pre-mRNA splicing of ERRγ. ERRγ increases FAO via regulating CPT1B [298]. FTO demethylates the transcription factors c-Jun, JunB, C/EBPβ, and c-Myc, thus rewiring glycolytic metabolism [299]. In LUAD, decreased FTO upregulates m6A abundance on MYC mRNA and enhances glycolysis [300]. In bladder cancer, decreased AlkB homolog 5 RNA demethylase (ALKBH5) promotes glycolysis by stabilizing CK2α mRNA in an m6A-dependent manner [301]. In metastatic renal cell carcinoma (RCC), downregulated methyltransferase-like 14 (METTL14) reduces m6A levels and stabilizes BPTF, which alters the super-enhancer landscape, affects DNA accessibility, and promotes glycolytic reprogramming [302]. YTHDF2 mediates m6A-dependent mRNA decay of LXRA, which is involved in cholesterol homeostasis control [303].
M5C RNA modification can bridge transcription and translation. The m5C modification on PKM2 mRNA can be recognized and stabilized by Aly/REF nuclear export factor (ALYREF) to facilitate glycolysis and cell proliferation [304]. Similar to DNA and histone modifications, RNA modifications regulate cancer metabolism, and conversely, cancer-specific metabolic changes can affect RNA modifications. RNA-modifying enzymes are potential therapeutic targets for cancer therapy [305]. FB23-2, a newly developed FTO inhibitor, can inhibit proliferation, promote differentiation, and induce apoptosis in AML cells, showing efficacy in treating AML [306]. However, there are no currently available small-molecule activators or inhibitors that selectively target RNA methyltransferases. Although the development of targeted drugs is still in a very early stage, their clinical applications might be very promising.
7 THERAPEUTIC PROSPECTS AND CLINICAL TRANSFORMATION
Previous clinical trials have suggested that using a single epigenetic or metabolic agent is insufficient. Based on the topic of this review, it is interesting to test whether metabolic or epigenetic abnormalities sensitize tumor cells to other epigenetic drugs, metabolic agents, or combined therapies. The aforementioned studies have provided a source of inspiration for identifying novel targets.
7.1 Challenges of epigenetic and metabolic monotherapy
DNA and histone modifications are both highly dynamic and reversible. Small-molecule compounds can potentially reverse aberrant epigenetic modification patterns during tumorigenesis, some of which have been approved for clinical use in hematological malignancies [307]. However, the therapeutic effect of monotherapy is not satisfactory for all patients and lacks efficacy for other solid tumors [308]. This raises interest in using combinations of epigenetic therapies with other agents in chemotherapies, immunotherapies, or targeted therapies to achieve synergistic effects. Analogously, despite many drugs targeting cancer metabolism entering clinical trials, few metabolic therapies have been approved [309, 310]. Metabolic heterogeneity and plasticity may account for the failed applications [311]. Therefore, it is necessary to identify bona fide metabolic vulnerability in a certain type of cancer. Metabolic alterations have also been found to be involved in treatment resistance. The combined use of metabolic agents may unlock the potential of epigenetic drugs and provide new clinical opportunities [312]. There are some possible ways to identify novel targets. First, basic researches have employed transcriptomics, epigenomics, and metabolomics to discover many new potential targets. For example, analysis of the metabolome of tumor cells after epigenetic agent GSK126 treatment reveals that lipid synthesis is strengthened to mediate drug resistance. Thus, targeting lipid metabolism can restore sensitivity to epigenetic therapy [313]. Second, combination drug screens with selected drug libraries targeting the metabolic and epigenetic abnormalities exhibited in tumors may provide more direct evidence to develop optimal therapies [314]. Third, current clinically proven treatment strategies may be extended to other cancer types possessing similar metabolic and epigenetic abnormalities. Testing these strategies will offer new therapeutic options for tumors that lack effective treatments.
7.2 Metabolic agents support antitumor effects of epigenetic therapy
Recently, the therapeutic potential of epigenetic agents in combination with metabolic inhibitors has attracted considerable attention. For IDH1-mutant AML, the mIDH1 (mutant IDH1) inhibitor ivosidenib and the hypomethylating agent azacitidine showed promising therapeutic effects in both preclinical stages and clinical trials. Encouraging results from a phase 3 trial showed that patients treated with ivosidenib and azacitidine combined therapy experienced greater clinical benefits than those treated with azacitidine monotherapy [204, 205, 315]. These works remind us of other cancer types with similar mutational and epigenetic patterns, such as glioma, sarcoma, and cholangiocarcinoma [316, 317]. Several small-molecule inhibitors targeting the glioma epigenome, such as mIDH inhibitors, HDAC inhibitors and DNMT inhibitors, are under clinical evaluation. A new clinical trial is underway to examine the effect of the combination of olutasidenib (mIDH1 inhibitor) with azacitidine in advanced glioma and chondrosarcoma [318].
Another breakthrough was discovering the potent synergistic anticancer effect of hypomethylating agents and BCL2 inhibitor venetoclax in solid tumors and hematological malignancies. Epigenetic drugs that inhibit DNMT, HDAC, and HMT trigger a marked metabolic shift from glycolysis to OXPHOS, which could generate excessive oxidative stress. Venetoclax then boosts the apoptosis of tumor cells by depolarizing the mitochondrial membrane and disrupting mitochondrial metabolism [276, 319, 320]. These drug combinations deliver a powerful one-two punch to cancer cells and have been successfully translated into clinical trials on leukemia and myelodysplastic syndrome [277, 321]. More importantly, solid tumors, such as liver, lung, colon and breast cancer, synergistically respond to these drug combinations. Further studies are necessary to determine whether their extraordinary results will be recapitulated.
Clinical experience suggests that epi-drugs are often ineffective in solid tumors, restricting their further applications. Thus, unraveling the underlying mechanisms of drug resistance or insensitivity is urgently needed. Epigenetic agents may also induce specific metabolic vulnerabilities in solid tumors, which could be exploited to develop innovative combinatorial treatment regimens. The EZH2 inhibitor GSK126 could change the overall metabolic profiles of melanoma, as evidenced by enhanced lipid synthesis. Drugs targeting fatty acid metabolism can re-sensitize tumor cells to EZH2 inhibition [313]. In cervical cancer, inhibition of HDAC makes cancer cells rely on glucose and glutamine catabolism for survival. Glycolysis and glutamine metabolism blockers, combined with HDAC inhibitors, further induce oxidative and energetic stress, accelerating cancer cell apoptosis [322, 323]. In glioblastoma, HDAC inhibitors elicit profound metabolic changes characterized by enhanced FAO but a decreased Warburg effect. The interaction and cause-and-effect relationship between epigenetic and metabolic processes provide a rationale for the combined use of the pan-HDAC inhibitor panobinostat and FAO inhibitor etomoxir. Combination treatment has shown better therapeutic effects than any single agent in patient-derived xenograft models [324] (Table 2).
| Study type | Potential therapeutic target | Agent | Cancer type | Investigation |
|---|---|---|---|---|
| Phase 3 (NCT03173248) [204, 205] | DNMT and mIDH1 | Azacitidine combined with ivosidenib | Leukemia | Efficacy and safety in treating IDH1m AML patients not suitable for standard induction therapy |
| Phase 1b/2 (NCT02677922) [315, 338] | DNMT and mIDH2 | Azacitidine combined with enasidenib | Leukemia | Efficacy and safety in IDH2m AML patients not suitable for standard induction therapy |
| Phase 1/2 (NCT03684811) [339] | DNMT and mIDH1 | Azacitidine combined with olutasidenib | Glioma and chondrosarcoma | Efficacy and safety in treating IDHm patients with advanced solid tumors and gliomas |
| Phase 3 (NCT02993523) [276, 277] | DNMT and BCL2 | Azacitidine combined with venetoclax | Leukemia | Efficacy and safety in treating AML patients not suitable for standard induction therapy |
| Phase 2 (NCT03404193) [340, 341] | DNMT and BCL2 | Decitabine combined with venetoclax | Leukemia | Efficacy and safety in treating patients with relapsed/refractory AML or relapsed high-risk Myelodysplastic Syndrome (MDS) |
| Phase 2 (NCT05376111) | DNMT and BCL2 | Azacitidine combined with venetoclax | Leukemia | Efficacy and safety in treating newly diagnosed T-cell acute lymphoblastic leukemia patients |
| Phase 2 (NCT03573024) | DNMT and BCL2 | Azacitidine combined with venetoclax | Leukemia | Efficacy and safety in treating non-elderly adult AML patients |
| Phase 2 (NCT04062266) | DNMT and BCL2 | Azacitidine combined with venetoclax | Leukemia | Efficacy and safety in treating patients with AML in remission |
| Phase 2 (NCT05361057) | DNMT and BCL2 | Azacitidine combined with venetoclax | Leukemia | Efficacy and safety in preventing relapse of consecutive measurable residual disease positive AML patients |
| Phase 2 (NCT04905810) | DNMT and BCL2 | Azacitidine or decitabine combined with venetoclax | Leukemia | Efficacy and safety in treating AML patients with prior hypomethylating agent failure |
| Phase 2 (NCT04801797) | DNMT and BCL2 | Azacitidine combined with venetoclax | Leukemia | Efficacy and safety in treating newly diagnosed AML |
| Phase 2 (NCT05048615) | DNMT and BCL2 | Azacitidine combined with venetoclax | Leukemia | Efficacy and safety in treating AML patients not suitable for intensive chemotherapy |
| Phase 2 (NCT05431257) | DNMT and BCL2 | Azacitidine combined with venetoclax | Leukemia | Efficacy and safety in treating elderly/unfit for standard therapy and relapsed/refractory patients with AML |
| Phase 2 (NCT04867928) | DNMT and BCL2 | Azacitidine combined with venetoclax | Leukemia | Efficacy and safety in managing molecular relapse/progression in NPM1-mutated AML patients |
| Phase 2 (NCT04128501) | DNMT and BCL2 | Azacitidine combined with venetoclax | Leukemia | Efficacy and safety in treating post-transplant AML patients |
| Phase 3 (NCT04102020) [342] | DNMT and BCL2 | Azacitidine combined with venetoclax | Leukemia | Efficacy and safety in treating adult AML participants in the first remission after conventional chemotherapy |
| Phase 1/2 (NCT04550442) | DNMT and BCL2 | Azacitidine combined with venetoclax | Leukemia and MDS | Efficacy and safety in treating relapsed/ refractory high-risk MDS or chronic myelomonocytic leukemia |
| Phase 1b (NCT02966782) | DNMT and BCL2 | Azacitidine combined with venetoclax | MDS | Efficacy and safety in treating patients with relapsed/refractory MDS |
| Phase 1 (NCT02942290) | DNMT and BCL2 | Azacitidine combined with venetoclax | MDS | Efficacy and safety in treating patients with treatment-naïve higher-risk MDS |
| Phase 1/2 (NCT04160052) [343] | DNMT and BCL2 | Azacitidine combined with venetoclax | MDS | Efficacy and safety in treating patients with high-risk recurrent or refractory MDS |
| Phase 2 (NCT05379166) | DNMT and BCL2 | Azacitidine combined with venetoclax | MDS | Efficacy and safety in treating therapy-related or secondary MDS |
| Preclinical [114] | BRD4 and AMPK | JQ-1 combined with compound C | Leukemia | / |
| Preclinical [313] | EZH2 and fatty acid synthesis | GSK126 combined with fenofibrate | Melanoma | / |
| Preclinical [322] | HDAC and glucose/glutamine metabolism | LMK235 combined with 2-DG/BPTES | Cervical cancer | / |
| Preclinical [323] | HDAC and glycolysis | LAQ824 combined with 2-DG | Glioblastoma | / |
| Preclinical [324] | HDAC and fatty acids oxidation | Panobinostat combined with etomoxir | Glioblastoma | / |
- Abbreviations: DNMT, DNA methyltransferase; mIDH1, mutant isocitrate dehydrogenase 1; AML, Acute myeloid leukemia; mIDH2, mutant isocitrate dehydrogenase 2; BCL2, BCL2 apoptosis regulator; MDS, Myelodysplastic syndrome; BRD4, Bromodomain containing 4; AMPK, AMP-activated protein kinase; EZH2, Enhancer of zeste homolog 2; HDAC, Histone deacetylase.
7.3 Synthetic lethality principle in epigenetic-metabolic circuit
The concept of synthetic lethality can be summarized as the interaction between two genes. Loss of either gene alone does not affect cell viability, but the loss of both genes simultaneously leads to cell death [325, 326]. In other words, losing one of the two genes renders tumor cells highly dependent on another. Consequently, targeting the synthetic lethal partner is a potent anticancer strategy for oncogenic mutations previously thought to be pharmacologically intractable [327, 328]. One of the most classic examples of synthetic lethal interactions in cancers is the BRCA mutation and PARP inhibition [329, 330]. Since then, many other novel synthetic lethal interactions have been identified [328]. Loss-of-function mutations lack targeted therapeutic approaches, and some are vulnerable to metabolic inhibitors or epigenetic agents. Available evidence demonstrates that synthetic lethal screening is a promising therapeutic option for patients with epigenetic or metabolic deficiencies.
Cancer cells with epigenetic defects exhibit metabolic vulnerabilities. Recent findings have extended the synthetic lethal partners to proteins closely related to metabolism. BCAT1 inhibitors impair the proliferation of EZH2-deficient leukemia-initiating cells both in vitro and in vivo. Inhibition is selective and does not affect normal HPSCs and hematopoiesis. Inhibition of metabolism may also be applied to other types of hematological malignancies with EZH2 mutations or dysregulation [212]. In LUAD, KMT2D loss abolishes the inhibitory effect of PER2 on glycolytic genes. Increased glycolytic activity is an attractive therapeutic vulnerability. 2-DG preferentially hampers LUAD cell growth and tumor formation in xenotransplantation models [213]. TET3-depleted AML cells are sensitive to inhibition of glycolysis by 2-DG [207]. Lung cancer with SMARCA4 or ARID1A loss is characterized by enhanced OXPHOS. Extreme reliance on energy production makes SWI/SNF-mutant LUAD more susceptible to the OXPHOS inhibitor IACS-010759 than tumor cells without the aforementioned mutations [19]. ARID1A inactivation was synthetically lethal with GLS and GCLC inhibition. The loss of ARID1A leads to a metabolic phenotype characterized by glutamine dependence. ARID1A-mutant ovarian clear cell carcinoma cell lines and tumors formed in orthotopic xenograft models are sensitive to GLS inhibitor CB-839 [226]. Another study on ovarian carcinoma cells reported that ARID1A mutations have synthetic lethal relationships with the glutathione metabolic pathway. Pharmacologically inhibiting the key enzyme GCLC with buthionine-sulfoximine selectively induces ARID1A-deficient cancer cell death. Surprisingly, both the genetic and pharmacological inhibition of glutamine transport and catabolism are ineffective in cells with ARID1A deficiency [227].
Metabolic deficiencies create specific vulnerabilities to epigenetic agents. LKB1-mutant pancreatic tumor cells are susceptible to inhibition of the serine metabolic pathway and DNA methylation, which is the major consumer of SAM. DNMT inhibitor decitabine hinders tumor growth, induces necrosis and apoptosis, and causes significant tumor regression in vitro and in vivo [135]. Similarly, the loss of PKCλ/ι induces NEPC differentiation by controlling global DNA methylation levels. Decitabine blocks NEPC differentiation and inhibits tumor proliferation [138]. A shortage of glutamine leads to histone hypermethylation on H3K27, which helps melanoma cells develop drug resistance to BRAF inhibitors. However, abnormal histone methylation patterns confer crucial vulnerability to histone methyltransferase EZH2 inhibitors. DZNep and EPZ005687 inhibit tumor growth when combined with BRAF inhibitors to overcome tumor drug resistance [151] (Table 3).
| Epigenetic/metabolic defect | Cancer type | Potential therapeutic target | Agent | Study type |
|---|---|---|---|---|
| Epigenetic defect | ||||
| EZH2 deficiency [212] | Leukemia | BCAT1 | Gabapentin | Preclinical |
| KMT2D deficiency [213] | Lung cancer | Glycolysis | 2-DG | Preclinical |
| ARID1A deficiency [226] | Ovarian cancer | GLS | CB-839 | Preclinical |
| ARID1A deficiency [227] | Ovarian cancer | GCLC | Buthionine-sulfoximine | Preclinical |
| SMARCA4 deficiency [19] | Lung cancer | Oxidative phosphorylation | IACS-010759 | Preclinical |
| Metabolic defect | ||||
| LKB1 deficiency [135] | Pancreatic cancer | DNMT | Decitabine | Preclinical |
| PKCλ/ι deficiency [138] | Prostatic cancer | DNMT | Decitabine | Preclinical |
| ACLY overexpression [116] | Pancreatic cancer | BET and mevalonate pathway | JQ-1 and atorvastatin | Preclinical |
- Abbreviations: EZH2, Enhancer of zeste homolog 2; BCAT1, Branched-chain amino acid transaminase 1; KMT2D, Histone lysine methyltransferase 2D; ARID1A, AT-rich interacting domain-containing protein 1A; GLS, Glutaminase; GCLC, Glutamate-cysteine ligase synthetase catalytic subunit; SMARCA4, SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 4; LKB1, Liver kinase B1; DNMT, DNA methyltransferase; PKCλ/ι, Protein kinase C λ/ι; ACLY, ATP-citrate lyase; BET, Bromodomain and extra-terminal domain protein.
Suppressing a broad spectrum of metabolic or epigenetic enzymes can cause potential deleterious side effects. New-generation epigenetic drugs, such as BET inhibitors, HMT inhibitors, and KDM inhibitors, are more specific. Their applications may improve the efficacy and tolerability of synthetic lethal therapies and epigenetic drugs in combination with metabolic therapies.
8 CONCLUSIONS AND PERSPECTIVES
Cell metabolism and the epigenetic landscape are highly dynamic. Epigenetic abnormalities deregulate metabolic enzymes or signaling pathways to provide energy, nucleotides, amino acids, fatty acids and many other metabolites to cancer cells and support their rapid proliferation. Furthermore, nutritional status and intracellular signals coordinate gene expression at the epigenetic level by churning metabolite pools. These two cooperate to enable cancer cells to quickly adjust to the changing environment.
However, it is intriguing that the mutual regulation of metabolism and epigenetics is precise to some extent. Specifically, only limited and certain types of histone methylation are influenced when the intracellular SAM content fluctuates. It can be surmised that this phenomenon is ascribed to the different catalytic properties of the enzymes responsible for those methylation sites. Furthermore, not all metabolic pathways are selectively modulated by epigenetic lesions in cancer cells. KMT2D-mutated cancer cells consistently showed a dependency on glycolysis. In contrast, different cancers with the same epigenetic lesion as ARID1A inactivation tend to expose distinct metabolic fragilities. The mechanisms underlying these discrepancies warrant further investigation. It is worth noting that concomitant changes in diverse cellular processes occur inextricably when cellular metabolic states shift. For example, the AMPK and mTOR pathways are intrinsic metabolic sensors that monitor intracellular energy production and nutrient supply, controlling cell growth, proliferation, and survival [331]. In addition to being a methyl donor, increased availability of SAM could function as a signal molecule sensed by the SAM sensor upstream of mTORC1 (SAMTOR) and abrogate inhibition of the mTOR pathway [332]. Furthermore, as mentioned above, many enzymes share the same substrates or cofactors. They are also affected by metabolic disturbances and chromatin modifiers. For instance, oncometabolites can drive tumorigenesis by hampering the activity of prolyl-hydroxylases, which fosters the stabilization of HIF-1α, in addition to demethylases [333]. In addition, nonhistone proteins are widely modulated by various metabolite-induced post-translational modifications, affecting almost all aspects of cell biology, such as gene transcription and signal transduction. One case is p53, whose acetylation and methylation can fine-tune its transcriptional activity [334]. These extensive and unexpected biological effects on cancer may obfuscate the contribution of epigenetic mechanisms and require careful dissection.
Considering the highly intertwined relationship between metabolism and epigenetic regulation, it is not surprising that metabolic drugs can reverse epigenetic alterations, and in turn, epigenetic agents can exert antitumor effects partly by disturbing cancer metabolism [221]. High-throughput technologies will help characterize the specific epigenetic or metabolic vulnerabilities exposed during this two-way communication, which could be induced and exploited as potential therapeutic targets. Combined pharmacological intervention and synthetic lethal screening are feasible approaches. In particular, elegant studies combining metabolic therapy and epigenetic therapy in hematological malignancies provide a milestone in targeting the epigenetic-metabolic circuit, hopefully becoming a novel paradigm for cancer treatment. Although the prospect is exciting, most of our related knowledge is limited to in vitro studies and is usually context-specific, without considering the effects on immune cells [335-337]. More confirmatory evidence should be explored before actual clinical practice.
Recently, the burgeoning fields of ncRNAs and RNA epigenetics have provided novel insights into the crosstalk between epigenetics and cancer metabolism, the therapeutic values of which have not yet been comprehensively studied. Nonetheless, they can be regarded as candidate targets for developing new therapies.
In conclusion, this review highlights the close connections between metabolism and epigenetics in cancer and proposes promising targeting therapeutic strategies. The current preclinical and clinical studies knowledge will potentially open up further research and novel therapeutic opportunities.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
FUNDING
This work was supported by the National Natural Science Foundation of China (81600766), the Science and Technology Commission of Shanghai (20DZ2270800), and the Innovative research team of high-level local universities in Shanghai (SHSMU-ZDCX20210900; SHSMU-ZDCX20210902).
AUTHOR CONTRIBUTIONS
Xianqun Fan, Shengfang Ge, and Ai Zhuang designed and revised the manuscript. Tongxin Ge, Ai Zhuang, and Peiwei Chai wrote the manuscript and made the figures. Peiwei Chai, Xiang Gu, and Renbing Jia polished the manuscript and gave useful suggestions. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
The material supporting the conclusion of this review has been included in the article.




