Inhibition of glucuronidation in pancreatic cancer improves gemcitabine anticancer activity
Abbreviations
-
- Ac
-
- Agglomerative coefficient
-
- CCL
-
- ommercial cell lines
-
- CI
-
- Confidence interval
-
- DFS
-
- Disease-free survival
-
- FDR
-
- False discovery rate
-
- HR
-
- Hazard ratio
-
- HTS
-
- High transcriptomic similarity
-
- ICA
-
- Independent component analysis
-
- KRAS
-
- V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
-
- LTS
-
- Low transcriptomic similarity
-
- OS
-
- Overall survival
-
- NR
-
- Not reached
-
- PAMG
-
- Pancreatic adenocarcinoma molecular gradient
-
- PDAC
-
- Pancreatic ductal adenocarcinoma
-
- PDC
-
- Patient-derived primary cell cultures
-
- PDO
-
- Patient-derived organoids
-
- PDX
-
- Patient-derived xenografts
-
- PPI
-
- Protein-protein interactions
-
- SD
-
- Standard deviation
-
- TCGA
-
- The cancer genome atlas
-
- TP53
-
- Tumor protein 53
-
- UGT
-
- UDP-glucuronosyltransferase
-
- UGT1A
-
- UDP-glucuronosyltransferase 1A
-
- w/Gem
-
- with adjuvant gemcitabine
-
- wo/Gem
-
- without adjuvant gemcitabine
Dear Editor,
Pancreatic ductal adenocarcinoma (PDAC) treatment is focused on two regimens. The polychemotherapy, FOLFIRINOX (folinic acid, fluorouracil, irinotecan, oxaliplatin), is used in patients with good health conditions [1], while gemcitabine, as monotherapy, in patients with poor health conditions [2-4]. Gemcitabine resistance-associated pathways have been targeted to sensitize cancer cells, but the results were disappointing. Using a transcriptomic bioinformatics analysis combined with biological validation, we showed that glucuronidation was associated with the gemcitabine resistance in PDAC, and its inhibition could switch tumors from resistant to sensitive.
To unravel the biological drivers of gemcitabine response in PDAC, we determined the transcriptomic dissimilarity between two preclinical models with defined gemcitabine sensitivity (Supplementary Figure S1A). Hierarchical clustering was applied to the expression matrix of 90 patient-derived xenografts (PDXs) and 38 patient-derived primary cell cultures (PDCs). Seven patients with high transcriptomic similarity (HTS) between both preclinical models were detected (Figure 1A). This was confirmed by Spearman's correlation, where HTS samples displayed a higher coefficient than the low transcriptomic similarity (LTS) group (0.48 ± 0.11 vs 0.13 ± 0.18, P < 0.001, Figure 1B). Next, we defined a transcriptomic component that captured the biological mechanisms associated with gemcitabine response in the HTS group using independent component analysis (ICA). The selected component had to meet two conditions: (1) be able to capture PDC gemcitabine response and (2) have a homologous component into PDX biological latent spaces. ICA2 component displayed a high correlation with gemcitabine response in PDCs measured by the area under the curve (AUC, r = −0.93, P = 0.003, Figure 1C). Then, we determined the ICA2 inter-model stability into PDX biological components. ICA4 component extracted from the HTS-PDXs showed a high correlation with ICA2 (r = 0.93, P = 0.003, Figure 1D). The PDC-selected component was refined, reducing the gene number in intervals of 1 standard deviation until we achieved a higher correlation coefficient in the remaining 31 PDC samples (r = −0.43, P = 0.010, Figure 1E). This resulted in a reduced ICA2 component with 99 high contributive genes (GemCore). The GemCore score was validated in an independent cohort of 13 PDX samples (r = −0.63, P = 0.010, Figure 1E), 14 patient-derived organoids (PDOs, r = −0.70, P = 0.003, Figure 1E), and 11 commercial cell lines (CCL, r = −0.61, P = 0.027, Figure 1E).
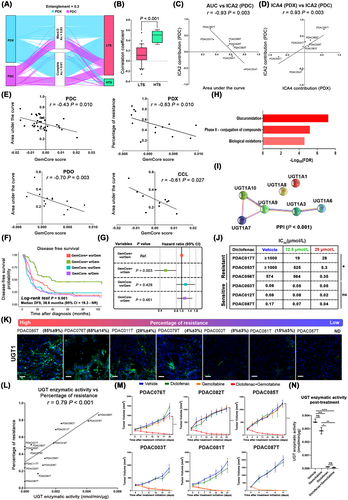
Derivation and validation of a gemcitabine-response component. (A). PDX and PDC agglomerative hierarchical clustering of transcriptomic data unravel 7 patients with high transcriptomic similarity (HTS, PDAC028T, PDAC031T, PDAC054T, PDAC083T, PDAC087T, PDAC089T and PDAC091T). (B) Boxplot of Spearman correlation coefficients shows a high correlation between PDXs and the corresponding PDCs in the HTS patients (0.48 ± 0.11) compared with the low transcriptomic similarity group (LTS, n = 31), which displayed a coefficient of 0.13 ± 0.18 (P < 0.001). (C) Identification of ICA2 component as gemcitabine-response component by independent component analysis on the 7 HTS PDCs and Spearman correlation with the AUC as selection parameter (r = −0.93, P = 0.003). (D) ICA2 sample contribution correlates positively with the ICA4 component extracted from the 7 HTS PDXs (r = 0.93, P = 0.003). (E) Scatterplots representing the correlation between the projected ICA2 and the response to gemcitabine measured by AUC, with Spearman test in an independent cohort of 31 PDCs (r = −0.43, P = 0.010), 13 PDXs (r = −0.63, P = 0.010), 14 PDOs (r = −0.70, P = 0.003) and 11 CCLs (r = −0.61, P = 0.027). (F) Nicole et al., cohort (n = 305) showed a significant patient stratification following the GemCore signature for disease-free survival (DFS). GemCore+ patients treated with gemcitabine showed a median DSF (38.6 months, 95% CI = 19.93-Not reached) higher than other groups (G) Nicolle et al. cohort GemCore+ patients treated with gemcitabine showed a DFS hazard ratio of 0.5 (95% CI = 0.32-0.79, P = 0.003). (H) GemCore pathway enrichment analysis shows an association between drug detoxification mechanisms, such as glucuronidation (FDR < 0.001), phase II conjugation of compounds (FDR < 0.001), and biological oxidations (FDR < 0.001) with gemcitabine resistance. (I) GemCore String enrichment analysis shows significant protein-protein interactions (PPI) between the UGT proteins (J) The IC50 after the treatment with diclofenac, observing a significant reduction in the IC50 in the resistant PDCs at 25 μmol/L. (K) UGT1A expression measured by immunofluorescence in a set of PDX samples distributed along the gemcitabine response profile, displaying a range from 85% to no detectable number of cells. Scale bar: 200 μm. (L) Spearman correlation of UGT activity and percentage of resistance in a set of fifteen PDXs (r = 0.79, P < 0.001). (M) Tumor growth curves for six PDXs (3 resistant and 3 sensitive) treated with vehicle, gemcitabine, diclofenac, or diclofenac plus gemcitabine. Resistant PDXs (PDAC076T, PDAC082T, and PDAC085T) treated with diclofenac and gemcitabine show a significant reduction in tumor volume compared with gemcitabine alone. (N) Box plot of UGT activity levels in resistant PDXs at the end time-point. *P < 0.05; **P < 0.01; ***P < 0.001. Results are shown as means ± SD. Differences between two groups are calculated by the Mann-Whitney test. Multi-group comparison is calculated by the Kruskal-Wallis test following Dunn's test
Additionally, GemCore was validated in two external cohorts of patients: (1) a subset of 80 patients treated with gemcitabine as an adjuvant from the cancer genome atlas (TCGA) and (2) 305 patients from the study by Nicolle et al. [5] where 170 patients were treated with adjuvant gemcitabine and 135 without a reported treatment. GemCore+ discriminated the higher survival of patients in the gemcitabine-treated group in the TCGA (Log-rank test P = 0.035 Supplementary Figure S1B) and Nicolle et al. cohorts (Log-rank test P = 0.002, Supplementary Figure S1C). The median overall survival (OS) of the TCGA GemCore+ patients was 24.6 months [95% confidence interval (CI) = 21.7 months-not reached], while in the Nicolle et al. cohort the median OS was not reached (95% CI = 50.6 months-not reached) for the GemCore+ patients treated with gemcitabine. The median disease-free survival (DFS) of the GemCore+ patients who received gemcitabine in the Nicolle et al. cohort (38.6 months, 95% CI = 19.3 months-not reached) was higher than the other groups (P = 0.001, Figure 1F). Cox proportional hazard model for overall survival showed a strong interaction between the GemCore stratification and gemcitabine treatment. TCGA GemCore- patients displayed a hazard ratio (HR) of 2 (95% CI = 1.10-3.90, P = 0.035, Supplementary Figure S1D), and in the Nicolle et al. cohort GemCore+ patients treated with gemcitabine showed a HR for OS of 0.45 (95% CI = 0.27-0.73, P = 0.001, Supplementary Figure S1E) and for DFS of 0.5 (95% CI = 0.32-0.79, P = 0.003, Figure 1G). In contrast with previous reports [5-7], GemCore+ patients were detected in both classical and basal-like subtypes and were distributed across the pancreatic adenocarcinoma molecular gradient (PAMG) in TCGA (Supplementary Figure S1F-G) and Nicolle et al. cohorts (Supplementary Figure S1H-I). No association was found between the gemcitabine response and the mutational profile of KRAS and TP53 (Supplementary Table S1). To identify the biological mechanisms associated with the gemcitabine response, we analyzed the pathways related to the gemcitabine-resistant phenotype. Pathway enrichment analysis on the GemCore revealed a significant role of detoxification pathways (Figure 1H-I) as determinants of gemcitabine response. Among them, glucuronidation was the only pathway with potential targets able to be modulated (Supplementary Table S2).
UGT1A expression and enzymatic activity were measured in 14 PDCs by immunofluorescent and fluorometric assays, respectively. PDCs showed high intra- and inter-heterogeneity in the percentage of positive cells for the UGT1A family, ranging from 90% to not detectable (Supplementary Figure S2A). UGT1A enzymatic activity was correlated positively with the percentage of positive cells (r = 0.82, P < 0.001, Supplementary Figure S2B) and the gemcitabine response (r = 0.76, P < 0.001, Supplementary Figure S2C), suggesting an association between the UGT1A family and the PDC resistant profile. The association between UGT1A and gemcitabine resistance was evaluated in vitro using two specific UDP-glucuronosyltransferase (UGT) inhibitors, demethylzeylasteral [8] (T-96) and hecogenin [9]. Fifteen PDCs were assessed in a cotreatment setting with two non-cytotoxic concentrations for each UGT inhibitor (Supplementary Figure S2D). T-96 and hecogenin displayed a strong sensitization effect on gemcitabine at the highest concentration of the inhibitors (25 and 12.5 μmol/L, respectively), and this effect was negatively correlated with the PDC UGT activity for both T-96 (r = −0.80, P < 0.001, Supplementary Figure S3A) and hecogenin (r = −0.76, P < 0.001, Supplementary Figure S3B). Because T-96 and hecogenin are not used in the clinics, and their UGT inhibition effect is isoform-dependent, we studied the effect of diclofenac, an unspecific pan-UGT inhibitor broadly utilized clinically [10]. Diclofenac's sensitization effect on gemcitabine was assessed in a subset of six (3 resistant and 3 sensitive) PDCs. Diclofenac improved the gemcitabine cytotoxic activity in the resistant PDCs (P = 0.011) but not in the sensitive ones (Figure 1J, Supplementary Figure S3C). These in vitro observations were validated in PDXs, where the UGT activity was correlated with the percentage of positive cells (r = 0.96; P < 0.001, Figure 1K, Supplementary Figure S4) and response to gemcitabine (Figure 1L). We observed a reduction in tumor volume in the resistant PDXs co-treated with gemcitabine and diclofenac compared with gemcitabine alone (P < 0.001 at the endpoint, Figure 1M). The tumor volume reduction was associated with a decrease in the UGT enzymatic activity in the gemcitabine-resistant PDXs (Figure 1N).These in vitro and in vivo results demonstrated that UGT activity was associated with gemcitabine resistance in PDAC, promoting faster elimination of gemcitabine. Additionally, UGT inactivation switches PDAC gemcitabine response from resistant to sensitive. However, we cannot discard that the observed effect could be influenced by the diclofenac activity on cyclooxygenase-1 and -2, which are the main targets of this drug.
In conclusion, we found that glucuronidation was one of the main mechanisms involved in gemcitabine resistance in PDAC. Moreover, we proposed the combination of diclofenac and gemcitabine as a transferable clinical strategy to increase the gemcitabine anticancer activity in resistant PDAC.
AUTHOR CONTRIBUTIONS
ND and JI designed the study and obtained funding. NAF, AMA, BC, MB, OG, and JR performed the experiments and analyzed the data. NAF, AMA, EC, and JI constructed the figures and wrote the manuscript. All authors discussed the results and suggested revisions. All authors read and approved the final version of the manuscript. The corresponding author is responsible for all aspects of this work, including data analysis and presentation.
ACKNOWLEDGMENTS
Not applicable.
COMPETING INTERESTS
ND and JI are funders of PredictingMed (Marseille, France). MB is employed by PredictingMed. All remaining authors have declared no conflicts of interest.
FUNDING
This work is part of the national program tumor identity card funded and developed by the National League Against Cancer. This work was supported by the National Cancer Institute (Grants number 2018-078, 2019-037, and 2018-079), Canceropole Provence-Alpes-Côte d'Azur, Amidex Foundation and the national institute of health and medical research.
ETHICAL APPROVAL AND CONSENT TO PARTICIPATE
The study was approved by the local ethics committee (South Mediterranean Personal Protection Committee) following patient informed consent collection. The PaCaOmics study was registered at www.clinicaltrials.gov with registration number NCT01692873. PDAC samples were collected between January 2012 and December 2015. All experimental procedures on animals were approved by the Marseille Ethics Committee number 14 (C2EA-14).
CONSENT FOR PUBLICATION
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
TCGA cohort was downloaded with TCGA biolinks R package. Nicolle et al. cohort was requested directly to the authors, and CEL files from CCL were downloaded from the ArrayExpress database (E-MTAB-3610).




