Limitation and challenges in using pancreatic cancer-derived organoids as a preclinical tool
Abbreviations
-
- 5-FU
-
- 5-fluorouracil
-
- CDA
-
- cytidine deaminase
-
- HC-PAMG
-
- High contributive genes from the PDAC molecular gradient
-
- ICA
-
- Independent component analysis
-
- IGF1
-
- Insulin-like growth factor 1
-
- LGR5
-
- Leucine-Rich Repeat Containing G Protein-Coupled Receptor 5
-
- LGR6
-
- Leucine-Rich Repeat Containing G Protein-Coupled Receptor 6
-
- PDAC
-
- Pancreatic ductal adenocarcinoma
-
- PDC
-
- Patient-derived cell cultures
-
- PDO
-
- Patient-derived organoids
-
- RSPO1
-
- R-Spondin 1
-
- WNT3A
-
- Wingless-Type MMTV Integration Site Family, Member 3A
-
- WNT7B
-
- Wingless-Type MMTV Integration Site Family, Member 7B
Dear Editor,
Pancreatic ductal adenocarcinoma (PDAC) is a dismal disease with a fast evolution and unpredictable treatment response. Nowadays, FOLFIRINOX [1] and gemcitabine [2] are the preferred treatments with a response rate of 33% and 11%, respectively. This poor patient response has been associated with an inefficient/non-personalized treatment allocation. Consequently, developing a rapid and efficient preclinical tool to test tumor drug sensitivity for each patient is hugely needed. Biopsy patient-derived organoid (PDO) appears to be a promising tool for developing individualized treatments for patients with PDAC. Several PDO-based platforms are in development worldwide as a guide to optimize therapy by directing tailored treatments. A critical point to consider PDO as promising is that it must represent the great clinical heterogeneity of PDAC as much as possible. Moreover, PDO has displayed histological features that mimic the PDAC phenotype. These characteristics make PDO an interesting option to obtaining reliable chemo-response profiles at a reasonable timeframe for most PDAC patients. However, although PDO has potential advantages as a preclinical tool, several concerns related to their phenotype stability in culture have recently arisen. Specifically, PDO culture media contain several growth factors and small-molecule inhibitors, which induce phenotypic modifications from the original tumor that could be easily evaluated at the transcriptome level. This is a central point considering that PDAC drug sensitivity highly depends on the transcriptomic phenotype [3-6]. Therefore, is PDO a faithful model that reproduces in vitro the huge phenotypical heterogeneity observed in PDAC? Here, we analyzed the transcriptome of several PDAC preclinical models in association with the chemo-response profile for gemcitabine, 5-fluorouracil (5-FU), oxaliplatin and irinotecan. Furthermore, we characterized the pathways associated with PDO-specific phenotype.
Transcriptome dispersion was computed for PDO, patient-derived cell culture (PDC), patient-derived xenograft (PDX), and PDAC tumors using inter-sample Euclidean distance on the gene sets that define PurIST [5], the Chan-Seng-Yue classifier [7], and the high contributive genes from the PDAC molecular gradient (HC-PAMG) [6]. These three well-known stratification patterns are strongly associated with PDAC prognosis and drug response phenotype. PDO displayed the lowest transcriptomic dispersion compared with PDC, PDX, and PDAC tumors in all the tested gene sets (Figure 1A). Moreover, we observed that the low transcriptome variability of PDO was accompanied with an extreme enrichment in a classical phenotype (Figure 1B). On the contrary, PDC showed a strong displacement toward basal-like/basal A, whereas PDX was the most diverse and proximal to PDAC tumor displaying all the subtypes described by the Chan-Seng-Yue classifier (Figure 1B).
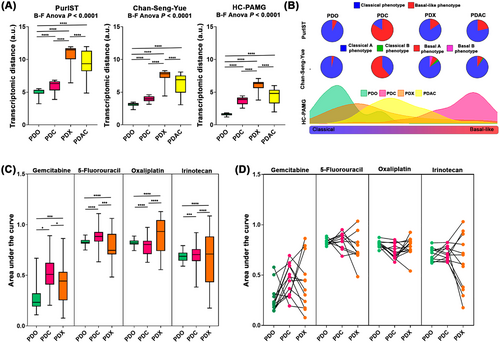
To further investigate the association between the transcriptome variability and the drug response profile among these preclinical models, 43 PDOs, 54 PDCs, and 18 PDXs were evaluated for gemcitabine, 5-FU, oxaliplatin, and irinotecan (Supplementary Figure S1A). A strong association between the degree of transcriptome dispersion and response profile was observed. PDO showed the lowest range of chemo-response measured by the area under the curve (AUC) for the four anti-cancer drugs, followed by PDC and PDX (Figure 1C). These observations were confirmed in a paired scheme where PDX was the preclinical model with the broadest response (Figure 1D).
Next, we analyzed the potential PDO phenotype drivers. Independent component analysis (ICA) was performed using PDAC molecular gradient calculated with HC-PAMG as a reference parameter. The ICA4 component showed the highest correlation with HC-PAMG (r = 0.57, P = 0.002, Supplementary Figure S1B). Pathway enrichment analysis on the ICA4 component revealed that Wnt signaling was a strong promoter of the PDO phenotype (Supplementary Figure S1C). Moreover, we identified that PDO had significantly higher WNT7B, LGR5, and LGR6 levels than PDC, PDX, and PDAC tumors (Supplementary Figure S1D). These observations were confirmed by differential expression analysis between PDO and PDAC tumors (Supplementary Figure S1E, Supplementary Table S1), where we detected activation of the Wnt signaling pathway and cell cycle in PDO compared to PDAC tumors (Supplementary Table S1). Additionally, we analyzed the expression of biomarkers associated with gemcitabine response, such as cytidine deaminase (CDA) [4] and insulin-like growth factor 1 (IGF1) [8]. CDA displayed a strong association with the gemcitabine response profile for the preclinical models and PDAC tumors, whereas IGF1 did not associate with gemcitabine response (Supplementary Figure S1F).
The findings provided by the current analysis validate the association between the transcriptomic variability and drug response in the PDAC preclinical models and highlight the limitations associated with the model settings. Specifically, we demonstrated that PDO displayed a reduced transcriptomic variability compared to PDC and PDX. This limited heterogeneity is reflected in the narrow chemo-response profile for the most common PDAC anti-cancer drugs. In addition, our results suggest a strong role of theWnt/R-spondin signaling as a driver of the PDO phenotype, which could be triggered by PDO culture conditions. These observations are aligned with recent studies that demonstrated a PDO-specific profile is highly divergent from the source PDAC biopsy [9]. A potential strategy to overcome the observed limitations of PDO is to modify the culture medium to capture PDAC heterogeneity, similar to the strategy proposed for breast cancer [10].
In conclusion, although the PDO is a promising tool for studying PDAC biology, we must consider the current limitations of this model for its use in a clinical setting, such as determining treatment protocol. Moreover, the lack of recapitulation of PDAC heterogeneity associated with a reduced response spectrum for the anti-cancer drugs indicates that further optimization is needed. Therefore, the culture conditions need to be improved to avoid a phenotypic polarization mediated by the over-activation of the Wnt/R-spondin pathways.
DECLARATIONS
ACKNOWLEDGMENTS
This work was supported by National Institute of Cancer (Grants number 2019-37, 2018-078 and 2018-079), Amidex Foundation and National Institute of Health and Medical Research.
CONFLICTS OF INTEREST
The authors declare no potential conflicts of interest
AUTHORS’ CONTRIBUTIONS
NAF and AMA: acquisition of data; analysis and interpretation of data; drafting of the manuscript; statistical analysis;
ND: study concept and design; analysis and interpretation of data; critical revision of the manuscript for important intellectual content; obtained funding; study supervision;
JI: study concept and design; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; obtained funding; study supervision.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was approved by the local ethics committee (Comité de protection des personnes Sud Méditerranée I) following patient informed consent collection. The PaCaOmics study was registered at www.clinicaltrials.gov with registration number NCT01692873. PDAC samples were collected between January 2012 and December 2015. All experimental procedures on animals were approved by the Comité d’éthique de Marseille numéro 14 (C2EA-14).
CONSENT FOR PUBLICATION
Not applicable.
DATA AVAILABILITY STATEMENT
ICGC-PACA-AU Seq expression dataset was downloaded from the International Cancer Genome Consortium (ICGC) data portal (https://dcc.icgc.org/). TCGA-PAAD was downloaded using The Cancer Genome Atlas (TCGA) biolinks R package. PDX dataset is available from ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) under the accession number E-MTAB-5039.




