KRAS-G12D mutation drives immune suppression and the primary resistance of anti-PD-1/PD-L1 immunotherapy in non-small cell lung cancer
Abstract
Background
Although immune checkpoint inhibitors (ICIs) against programmed cell death protein 1 (PD-1) and its ligand PD-L1 have demonstrated potency towards treating patients with non-small cell lung carcinoma (NSCLC), the potential association between Kirsten rat sarcoma viral oncogene homolog (KRAS) oncogene substitutions and the efficacy of ICIs remains unclear. In this study, we aimed to find point mutations in the KRAS gene resistant to ICIs and elucidate resistance mechanism.
Methods
The association between KRAS variant status and the efficacy of ICIs was explored with a clinical cohort (n = 74), and confirmed with a mouse model. In addition, the tumor immune microenvironment (TIME) of KRAS-mutant NSCLC, such as CD8+ tumor-infiltrating lymphocytes (TILs) and PD-L1 level, was investigated. Cell lines expressing classic KRAS substitutions were used to explore signaling pathway activation involved in the formation of TIME. Furthermore, interventions that improved TIME were developed to increase responsiveness to ICIs.
Results
We observed the inferior efficacy of ICIs in KRAS-G12D-mutant NSCLC. Based upon transcriptome data and immunostaining results from KRAS-mutant NSCLC, KRAS-G12D point mutation negatively correlated with PD-L1 level and secretion of chemokines CXCL10/CXCL11 that led to a decrease in CD8+ TILs, which in turn yielded an immunosuppressive TIME. The analysis of cell lines overexpressing classic KRAS substitutions further revealed that KRAS-G12D mutation suppressed PD-L1 level via the P70S6K/PI3K/AKT axis and reduced CXCL10/CXCL11 levels by down-regulating high mobility group protein A2 (HMGA2) level. Notably, paclitaxel, a chemotherapeutic agent, upregulated HMGA2 level, and in turn, stimulated the secretion of CXCL10/CXCL11. Moreover, PD-L1 blockade combined with paclitaxel significantly suppressed tumor growth compared with PD-L1 inhibitor monotherapy in a mouse model with KRAS-G12D-mutant lung adenocarcinoma. Further analyses revealed that the combined treatment significantly enhanced the recruitment of CD8+ TILs via the up-regulation of CXCL10/CXCL11 levels. Results of clinical study also revealed the superior efficacy of chemo-immunotherapy in patients with KRAS-G12D-mutant NSCLC compared with ICI monotherapy.
Conclusions
Our study elucidated the molecular mechanism by which KRAS-G12D mutation drives immunosuppression and enhances resistance of ICIs in NSCLC. Importantly, our findings demonstrate that ICIs in combination with chemotherapy may be more effective in patients with KRAS-G12D-mutant NSCLC.
List of abbreviations
-
- NSCLC
-
- non-small cell lung cancer
-
- LUAD
-
- lung adenocarcinoma
-
- LUSC
-
- lung squamous cell carcinoma
-
- ALK
-
- anaplastic lymphoma kinase
-
- EGFR
-
- epidermal growth factor receptor
-
- KRAS
-
- Kirsten rat sarcoma viral oncogene homolog
-
- ICIs
-
- immune checkpoint inhibitors
-
- PD-1
-
- programmed cell death protein 1
-
- PD-L1
-
- programmed cell death-ligand 1
-
- TIME
-
- tumor immune microenvironment
-
- TILs
-
- tumor-infiltrating lymphocytes
-
- TMB
-
- tumor mutational burden
-
- MAPK
-
- mitogen-activated protein kinase
-
- PI3K
-
- phosphatidylinositol 3-kinase
-
- CTLA-4
-
- cytotoxic T-lymphocyte antigen 4
-
- MSKCC
-
- Memorial Sloan Kettering Cancer Center
-
- CICAMS
-
- Cancer Hospital and Institute, Chinese Academy of Medical Sciences
-
- IHC
-
- immunohistochemistry
-
- BEGM
-
- bronchial epithelial cell growth medium
-
- FBS
-
- fetal bovine serum
-
- RT-qPCR
-
- reverse transcription quantitative PCR
-
- Erk1/2
-
- extracellular signal-regulated kinase 1 and 2
-
- HMGA2
-
- high mobility group protein A2
-
- CXCL10
-
- chemokine (C-X-C motif) ligand 10
-
- PBS
-
- phosphate buffer saline
-
- TPS
-
- tumor proportion score
-
- DAPI
-
- 4',6-diamidino-2-phenylindole
-
- ELISA
-
- enzyme linked immunosorbent assay
-
- PBMCs
-
- peripheral blood mononuclear cells
-
- PAM
-
- protospacer adjacent motif
-
- TCGA
-
- The Cancer Genome Atlas
-
- GO
-
- Gene ontology
-
- KEGG
-
- Kyoto Encyclopedia of Genes and Genomes
-
- TIMER
-
- Tumor Immune Estimation Resource
-
- PFS
-
- progression-free survival
-
- DCB
-
- durable clinical benefit
-
- mAb
-
- monoclonal antibodies
-
- GZMB
-
- Granzyme B
-
- mRNA
-
- messenger RNA
-
- mTOR
-
- mechanistic target of rapamycin
-
- PDO
-
- patient-derived organoid
-
- PD
-
- progressive disease
-
- OS
-
- overall survival
-
- TGF-beta
-
- transcription of growth factor beta
-
- EMT
-
- epithelial-to-mesenchymal transition
1 INTRODUCTION
Lung cancer, poses a serious threat to human health, as indicated by the top-ranking morbidity and mortality rates worldwide [1] and in China [2, 3]. Non-small cell lung cancer (NSCLC), including lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), accounts for ∼85% of lung cancers in patients as the most common pathological subtype [4]. Nevertheless, more than 40% of those diagnosed with NSCLC are already at stage IIIB or IV, whereby the best opportunity for surgical removal of the tumor has been missed [5]. Benefiting from the rapid development of medical molecular biology in the past decade, the therapies for metastatic or advanced NSCLC have transitioned from cytotoxic chemotherapy into the era of molecular targeted therapy [6]. For example, tyrosine kinase inhibitors against anaplastic lymphoma kinase (ALK) and epidermal growth factor receptor (EGFR) have been extensively used as targeted therapies for NSCLC in the clinic [7-10]. In contrast, despite clinical studies on the small-molecule inhibitors AMG510 [11] and MRTX849 [12], targeting the Kirsten rat sarcoma viral oncogene homolog (KRAS) G12C mutation, most countries have no routinely used KRAS-targeted therapies [13].
In recent years, immune checkpoint inhibitors (ICIs) against programmed cell death protein 1 (PD-1) and its ligand programmed cell death-ligand 1 (PD-L1) have revolutionized the treatment paradigm of NSCLC and provided an approach for the treatment of refractory patients with NSCLC [14], particularly patients with KRAS mutations [15, 16]. Our previous publication [17] proved that patients with KRAS-mutant NSCLC have a superior response to ICIs due to an inflammatory tumor immune microenvironment (TIME) with adaptive immune resistance. However, only 58% of NSCLC patients with KRAS-mutant NSCLC were shown to have a high expression of PD-L1 and infiltration of CD8+ tumor-infiltrating lymphocytes (TILs), and not all patients benefited from anti-PD-1 immunotherapy. We speculated that the patients who presented with primary drug resistance might lack an inflammatory TIME. Therefore, exploration of the TIME in patients with KRAS-mutant NSCLC is needed to effectively distinguish responders from non-responders treated with ICIs. By changing the TIME, a greater proportion of the population could benefit from immunotherapy, thereby establishing a new treatment paradigm for patients with KRAS-mutant NSCLC.
KRAS is an important driver gene in the development of NSCLC, and the mutation rates of KRAS in Western [18] and Asian patients [19] with NSCLC are 20%-30% and 10%-15%, respectively. In NSCLC, KRAS oncogene substitutions often occur at codons 12 and 13 of exon 2, with common codon variants containing G12C (c.34G > T, 32.11%), G12D (c.35G > A, 23.39%), G12V (c.35G > T, 21.10%), and G12A (c.35G > C, 12.84%) [20, 21]. Of these substitutions, G12C, G12V and G12A are transversion mutations, while G12D is a transition mutation [22]. Notably, previous studies have shown that NSCLC with G12C, G12V, and G12A mutations are typical for smoking-related tumors with high tumor mutational burden (TMB), while the KRAS-G12D point mutation is an exception [23, 24]. In addition, several studies have shown that not all subtypes of KRAS mutations have similar biological effects. Subtype heterogeneity also exists with regards to the survival prognosis of patients [21] and the efficacy of chemotherapy [22], molecular targeted therapy [25] and immunotherapy [26]. As shown previously [27, 28], the GTPase activity of the KRAS-G12V point mutant protein is one-fourth that of the G12D point mutant protein, and one-tenth that of wild-type KRAS. Compared with the G12D point mutant protein, the G12V point mutant protein binds tighter to GTP. Consequently, the G12V point mutant protein exists in a persistent spontaneous activated state. Furthermore, in addition to the mitogen-activated protein kinase (MAPK) signaling pathway, different substitutions of the KRAS oncogene activate various downstream signaling pathways. For example, the G12C and G12V mutations enhance the RalGDS-Ral pathway, while the G12D point mutations activates the phosphatidylinositol 3-kinase (PI3K)-AKT pathway [27]. In part, these mechanisms explain the disparities among the subtypes between the prognostic value and malignant transformation ability. Some recent studies have focused on the association between mutant variants of the KRAS gene and tumor immunity-related characteristics, including TMB, PD-L1 expression, and the presence of immune cells. However, the underlying association between KRAS oncogene substitutions and the efficacy of immunotherapy against PD-1/PD-L1 remains unclear, as does the mechanism of different substitutions, leading to immunotherapeutic heterogeneity [23, 29, 30].
In this study, the relationship between KRAS variant status and the efficacy of ICIs was investigated using an online clinical cohort, as well as cellular and mouse models. The immune landscapes, including CD8+ TILs, PD-L1, and immune-related genes levels, of patients with NSCLC harboring different substitutions of KRAS oncogene were analyzed, and the pathway activation involved in the formation of TIME was explored to find interventions with improved responsiveness to immunotherapy targeting PD-1/PD-L1.
2 MATERIALS AND METHODS
2.1 Clinical cohorts
To analyze the association between the KRAS variant status and efficacy of anti-PD-1/PD-L1 immunotherapy in NSCLC, the clinical data of 240 patients treated with anti-PD-1/PD-L1 alone or in combination with anti-cytotoxic T-lymphocyte antigen 4 (anti-CTLA-4) immunotherapy between April 2011 and January 2017 at the Memorial Sloan Kettering Cancer Center (MSKCC, New York, NY, USA) was downloaded from a previously published clinical cohort study [31]. According to the inclusion criteria (patients had KRAS mutations and received anti-PD-1/PD-L1 monotherapy) and exclusion criteria (patients with EGFR mutation), the clinical data of 74 patients who had KRAS mutations and received anti-PD-1/PD-L1 monotherapy was included in the analysis.
To investigate the clinical outcome of patients with NSCLC harboring KRAS-G12D mutations after initiation of PD-1/PD-L1 blockade monotherapy or chemo-immunotherapy, the clinical information of 11 NSCLC patients who harbored KRAS-G12D mutations and received immunotherapy, with or without chemotherapy, at the Cancer Hospital and Institute, Chinese Academy of Medical Sciences (CICAMS, Beijing, China) was retrospectively collected between September 2017 and April 2021. The inclusion criteria were that patients had KRAS-G12D mutations and received ICIs; the exclusion criteria were that patients had EGFR mutation and received other treatments in addition to ICIs and chemotherapy. Of the 11 patients enrolled, 3 received anti-PD-1 agent monotherapy, and 8 received chemo-immunotherapy. The last follow-up time was June 15th, 2021.
A total of 112 KRAS-mutant NSCLC specimens surgically resected between January 2008 and December 2013 were collected from the biobank of CICAMS for immunohistochemistry (IHC) analysis. The inclusion criteria were that the tumor tissues were pathologically diagnosed as NSCLC with KRAS mutations; the exclusion criteria were that patients had EGFR mutation and received chemotherapy or radiotherapy before surgery. This study was approved by the Ethics Committee of CICAMS (approval number: #20/242-2438). The tissue samples were obtained with written informed consent from each patient.
2.2 Cell lines and cell culture
Immortalized human bronchial epithelial cells (Beas-2B) were cultured in bronchial epithelial cell growth medium (BEGM) media (#CC-3170; Lonza, Basel, Switzerland) with 10% fetal bovine serum (FBS, #35-081-CV; Corning, New York, NY, USA). Human lung cancer cell lines (Calu3, H1703, H2030, H358, SK-LU-1, A427, H441, H292, A549, and H460) were cultured in RPMI-1640 media (#10-040-CV; Corning) containing 10% FBS and antibiotic mixture (100 U/mL penicillin and 100 μg/mL streptomycin, #15140-122; Gibco, Billings, MT, USA). H2009 and 293T cells were cultured in Dulbecco's modified eagle medium (DMEM) media (#10-013-CVR; Corning) containing 10% FBS and antibiotic mixture. LA795 cells (clones from the lungs of a 615 mouse with LUAD) were cultured in RPMI-1640 media containing 10% FBS and an antibiotic mixture.
2.3 Reverse transcription quantitative PCR (RT-qPCR), Western blotting, flow cytometry, IHC, and immunofluorescence
RT-qPCR, Western blotting, flow cytometry and IHC were performed as previously described [17, 32, 33]. Total RNA was extracted with TRIzol reagent (#15596; Life Technologies, Carlsbad, CA, USA). TransScript® II All-in-One First-Strand cDNA Synthesis SuperMix kit (#AH341-01; TransGen, Beijing, China) was used for reverse transcription. RT-qPCR was performed with TransStart® Top Green qPCR SuperMix (#AQ132; TransGen) on ABI 7900HT Real-Time PCR thermocycler (Life Technologies). The human and mouse PCR primers used in this study are shown in Supplementary Tables S1 and Supplementary Tables S2, respectively.
The primary antibodies for Western blotting used in this study are listed as follows: PD-L1 (#13684T; Cell Signaling Technology, Danvers, MA, USA), KRAS (#3339T; Cell Signaling Technology), KRAS G12D (#14429S; Cell Signaling Technology), β-Tubulin (#2128T; Cell Signaling Technology), Flag (#F3165; Sigma-Aldrich, Saint Louis, MO, USA), GAPDH (#ab8245; Abcam, Cambridge, UK), extracellular signal-regulated kinase 1 and 2 (Erk1/2, #4695; Cell Signaling Technology), Phospho-Erk1/2 (Thr202/Tyr204) (#4370; Cell Signaling Technology), AKT (#4691; Cell Signaling Technology), Phospho-AKT (Ser473) (#4060; Cell Signaling Technology), Phospho-AKT (Thr308) (#13038; Cell Signaling Technology), p70S6K (#2708; Cell Signaling Technology), phospho-p70S6K (Thr389) (#9234; Cell Signaling Technology), p70S6K (#66638-1-Ig; Proteintech, Rosemont, IL, USA), high mobility group protein A2 (HMGA2; #8179S; Cell Signaling Technology), chemokine (C-X-C motif) ligand 10 (CXCL10, #ab137018; Abcam), CXCL11 (#MAB672-SP; R&D, Minneapolis, MN, USA) and Actin (#66009-1-Ig; Proteintech). Briefly, cell protein lysates were electrophoresed using PAGE Gel Quick Preparation Kit (#8012011; Dakewe, Shenzhen, Guangdong, China) and transferred to PVDF membranes (#P2120-2; APPLYGEN, Beijing, China). After incubation with the primary antibodies at 4°C overnight, the membranes with HRP (horseradish peroxidase)-conjugated secondary antibodies at room temperature for 1 h. Finally, we visualized the protein bands using enhanced chemiluminescence.
The antibodies for flow cytometry used in this study included PD-L1 (APC, #329707; BioLegend, San Diego, CA, USA) and its isotype control (APC, #401210; BioLegend). Briefly, the antibodies were used to incubate cells at 4°C for 30 min. Phosphate buffer saline (PBS, #D8537; Sigma-Aldrich) was then used to wash and suspend cells for flow cytometric analysis.
The primary antibodies for IHC used in this study are listed as follows: PD-L1 (SP263 clone, #740-4907; Ventana Medical Systems, Oro Valley, AZ, USA), CD8 (#ZA-0508; Zsbio Tech, Beijing, China), HMGA2 (#8179S; Cell Signaling Technology), CXCL10 (#MAB2662-SP; R&D), CXCL11 (#MAB672-SP; R&D), PD-L1 (#ab238697; Abcam), CD8 (#98941; Cell Signaling Technology), CXCL10 (#10937-1-AP; Proteintech) and CXCL11 (#MAB572-SP; R&D). Briefly, the tumor tissue sections were incubated with these primary antibodies at 4°C overnight and then incubated with HRP-conjugated secondary antibodies at 25°C for 1 h after antigen retrieval. The PD-L1 tumor proportion score (TPS) and the proportion of CD8+ T cells were assessed according to the evaluation criteria of the previously published approach [17]. The IHC score was calculated using the following formula: IHC score = staining intensity × percentage of positive tumor cells × 100. There are four grades of staining intensity: no color staining was recorded as 0, pale yellow staining as 1, yellow staining as 2, and brown-yellow staining as 3 [34].
The primary antibodies for immunofluorescence used in this study included CD86 (#GB13585; Servicebio, Wuhan, Hubei, China) and CD11c (#GB11059; Servicebio). Briefly, the tumor tissue sections were incubated with these primary antibodies at 4°C overnight, and then stained with 4',6-diamidino-2-phenylindole (DAPI, #28718-90-3; Beyotime, Shanghai, China) to label nuclei for 15 min. Next, the sections were probed with secondary antibodies with fluorescence according to the primary antibodies used.
2.4 Cell supernatant extraction and enzyme-linked immunosorbent assay (ELISA)
Cell supernatant extraction and ELISA were performed as previously described [32]. The concentration of free CXCL10 and CXCL11 protein in extracted cell supernatants was detected by the Human CXCL10/IP-10 Quantikine ELISA Kit (#DIP100; R&D) and the Human CXCL11/I-TAC Quantikine ELISA Kit (#DCX110; R&D) according to the manufacturer's instructions.
2.5 In vitro tumoricidal activity assays
Human peripheral blood mononuclear cells (PBMCs) were obtained from peripheral blood provided by healthy donators using Ficoll Paque Plus density centrifugation (#17-1440-02; GE Healthcare, Cleveland, OH, USA). We then used the Human CD8+ T Cell Isolation kit (#130-096-495; Miltenyi Biotec, Bergisch Gladbach, Germany) to isolate CD8+ T cells from the PBMCs, and cultured the isolated CD8+ T cells in RPMI-1640 media containing 10% FBS, 200 U/mL interleukin-2 (IL-2, #200-02-50; PeproTech, Rocky Hill, NJ, USA) and an antibiotic mixture. Meanwhile, we added human anti-CD3/CD28 Dynabeads (#40203D; Thermo Fisher Scientific, Waltham, MA, USA) in the media to activate CD8+ T cells for 3 days based on the manufacturer's protocol. After A427, SK-LU-1, H2009, H358 and H441 cells adhered to the plate overnight, we co-cultured tumor cells and activated CD8+ T cells in a ratio of 1:5 for 48 h in the presence of isotype control or nivolumab (200 μg/mL, #A2002; Selleck, Shanghai, China). Next, we used the Cell Counting Kit-8 (#CK04; Dojindo, Kumamoto, Japan) to quantify living tumor cells after removing T cells and cellular debris.
2.6 Construction of KRAS-knockout cells
We first designed the single-guide RNA (3’-CTGAATTAGCTGTATCGTCA-5’) for mice and single-guide RNA (5’-CAATGAGGGACCAGTACATG-3’) for human using DeepCRISPR [35]. For single-guide RNA cloning, the SpCas9 targeting vector (#H11761; OBiO, Shanghai, China) was then digested with BsmBI (#ER0451; Thermo Fisher Scientific) and ligated with BsmBI-compatible annealed oligos (OBiO). We harvested lentiviruses using 293T cells as previously described [36]. Briefly, 293T cells were co-transfected with the SpCas9 targeting vector and packaging plasmid (pLP1, pLP2, and pLP/VSVG; OBiO) using Lipofectamine 3000 (#L3000015; Invitrogen, Carlsbad, CA, USA). We harvested the infectious lentiviruses at 48 h after transfection. LA795 and Beas-2B cells were then infected with harvested lentiviruses by the addition of 6 μg/mL polybrene (#P4505; Sigma-Aldrich), and single-cell clones were selected with mCherry using flow cytometry sorting. The knockout effect of KRAS in the infected cells was confirmed by Western blotting and Sanger sequencing after selection.
2.7 Construction of stable KRAS-overexpressing cells
For stable overexpression of exogenous KRAS plasmids, coding for wild-type KRAS (WT), mutant KRAS-Gly12Ala (G12A), mutant KRAS-Gly12Cys (G12C), mutant KRAS-Gly12Asp (G12D), or mutant KRAS-Gly12Val (G12V), cDNA was ligated into the lentiviral vector (mouse: #H11219; human: #GL122; OBiO). It is worth noting that the protospacer adjacent motif (PAM) region of single-guide RNA needs synonymous mutation in the cDNA. According to the above-mentioned methods of lentivirus production and infection, we gained the infected cells. Infected single-cell clones were then selected with EGFP using flow cytometry sorting. Western blotting was performed to determine the efficiency of the overexpression.
2.8 In vivo mouse experiments
Female mice (specific pathogen-free, 5-6 weeks old) were used for the animal experiments. The mice were purchased from the Institute of Hematology, Chinese Academy of Medical School (Tianjin, China) and housed at the Center of Experimental Animals of CICAMS. The Animal Care and Use Committee of CICAMS approved all procedures concerning in vivo mouse experiments (approval number: #NCC2020A163).
A total of 615 mice were subcutaneously implanted with murine LA795-derived xenografts that harbored different KRAS oncogene substitutions, including G12A, G12C, G12D and G12V, and treated with anti-PD-L1 monoclonal antibody (10 mg/kg three times weekly; Bio X Cell, West Lebanon, NH, USA) or paclitaxel (20 mg/kg twice weekly; CSPC, Shijiazhuang, Hebei, China) when the tumor volume reached ∼100 mm3. After the treatment intervention, all mice were euthanized by carbon dioxide asphyxiation to dissect the subcutaneous tumor for IHC analysis.
2.9 Transcriptome sequencing data, immune cell infiltration analysis and TMB calculation
HTseq-Counts data of 139 LUAD samples with KRAS mutations were extracted from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) and divided into two groups with or without KRAS-G12D mutations. We previously identified 1,584 genes that were differentially expressed between the two groups at the thresholds of P < 0.05 and log2(fold-change) > 1 using the “DESeq2” R package [37]. Finally, 39 out of 730 immune-related genes extracted from the nCounter PanCancer Immune Profiling Panel (NanoString) [38] were identified. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed to determine the biological function and pathway correlated with the 39 differentially expressed genes in the DAVID 6.8 (http://david.abcc.ncifcrf.gov).
The Tumor Immune Estimation Resource (TIMER; https://timer.cistrome.org ) was used to calculate the fractions of six intra-tumoral immune cells in each sample, according to the gene expression profile data. Notably, quantile-normalization method was used to standardize the gene expression profile data to eliminate the influence of confounding variables [39].
Somatic mutation data of LUAD were downloaded from the MSKCC dataset (https://www.cbioportal.org/) and TCGA database. By counting the total count of somatic mutations detected in the coding region, except silent mutations, TMB was calculated for each tumor sample.
2.10 Organoid culture
LUAD samples with KRAS-G12D mutations were obtained from the CICAMS and were transported directly to the laboratory after the surgery was performed. The patients provided informed consent. The study was approved by the Ethics Committee of the CICAMS (approval number: #20/242-2438). The tumor samples were washed twice with cold PBS and minced into smaller pieces using scissors. The protocol for establishing organoids was previously described [40]. Organoids were cultured in OrganoProTM Tumor Organoids Culture Media (#K2O-M-NSCLC; Ketu Tech, Beijing, China), and passaged at a 1:3 ratio every 2-3 weeks.
2.11 Statistical analysis
SPSS v21.0 (SPSS, Chicago, IL, USA) and Prism v8.0 (GraphPad, San Diego, CA, USA) was used for data analysis. The experimental data are expressed as the mean ± standard deviation, and the data analysis between groups was conducted with Mann-Whitney U tests, one-way analysis of variance, and Student's t-test. Pearson correlation analysis was used to assess the association between samples. We used the Kaplan-Meier method to generate survival curves, and performed comparisons between groups by log-rank tests. P values less than 0.05 were considered statistically significant. The graphical abstract was created with BioRender.com. Based on the ARRIVE1 guidelines, we have completed the reporting checklist for study.
3 RESULTS
3.1 KRAS-G12D mutation was correlated with the primary resistance of ICI monotherapy in NSCLC
A clinical cohort of 74 NSCLC patients that harbored KRAS mutations and received anti-PD-1/PD-L1 monotherapy was enrolled to explore the correlation between KRAS variant status and the clinical benefit of ICIs (Supplementary Table S3). Depending on the variant status of the KRAS oncogene, these patients were assigned to five subgroups: G12A (n = 7), G12C (n = 30), G12D (n = 11), G12V (n = 12), and others (n = 14). As shown in Figure 1A, there were significant differences among the Kaplan-Meier survival curves of these five subgroups (P = 0.044). Notably, the NSCLC patients with KRAS-G12D mutations had significantly shorter progression-free survival (PFS) than those with other point mutations (P = 0.006; Figure 1B). Although the effect of immunotherapy is slow when compared with conventional anti-tumor therapy, a durable clinical benefit (DCB, PFS > 6 months) can often be obtained [15]. Differences were found among groups when analyzing the DCB of patients with different substitutions (Figure 1C). Of note, the proportion of patients achieved DCB was lower in the G12D group than in the non-G12D group (P = 0.028; Figure 1D). By investigating the relationships between KRAS oncogene substitution and TMB in these patients, the results showed that patients with KRAS-G12D mutations exerted the lowest TMB (Supplementary Figure S1A-B). This finding was also verified using mutation data from the TCGA database (Supplementary Figure S1C).

To further confirm the clinical findings, a co-cultured system consisting of cytotoxic CD8+ T lymphocytes and tumor cells was established to evaluate the tumoricidal activity of CD8+ T cells cocultured with tumor cells harboring different KRAS oncogene substitutions (Supplementary Table S4). Western blotting and flow cytometry were firstly performed to detect the protein level of PD-L1 in Beas-2B cells and 11 NSCLC cell lines. The results showed that cell lines with KRAS-G12D mutation (A427 and SK-LU-1) had a lower level of PD-L1 than the other cell lines without KRAS-G12D mutation (Figure 1E-F). Of note, the tumoricidal activity of CD8+ T cells cocultured with the cell lines harboring non-G12D mutations increased significantly after nivolumab intervention; yet the cells carrying G12D mutations did not (Figure 1G).
In addition to the co-culture model, a mouse model was established to evaluate the efficacy of anti-PD-L1 drugs. We first knocked out the KRAS gene in LA795 cells without KRAS mutation, and then transferred lentivirus vectors coding four common KRAS oncogene substitutions (G12A, G12C, G12D and G12V) in KRAS-knockout LA795 cells (Supplementary Figure S2A-C). A previous study reported that KRAS mutations at different sites did not affect cell growth [27]. To further explore whether KRAS mutations at different sites affect the growth of subcutaneous tumors in mice, we subcutaneously transplanted LA795 cells carrying different KRAS substitutions into nude mice. It was found that there was no significant difference in the growth of subcutaneous tumors in these groups (Supplementary Figure S2D-E). Next, mice loaded with different KRAS point mutations were established by subcutaneously transplanting tumor xenografts derived from mouse LA795 cell lines carrying different KRAS substitutions into 615 mice (Figure 2A). Consistent with our findings in the clinical cohorts, anti-PD-L1 monoclonal antibodies (mAb) in mice with G12D mutations were less effective against tumor growth compared to other groups (P < 0.001; Figure 2B). Furthermore, from IHC analysis, there was a lower protein level of PD-L1 and less CD8+ T cell infiltration in mouse tumor specimens carrying the KRAS-G12D point mutation before the intervention of anti-PD-L1 antibodies compared to the other groups. In contrast, after anti-PD-L1 antibody intervention, the mice in the G12D group did not show a markedly decreased PD-L1 protein level and increased CD8+ T cell infiltration as did the other groups (Figure 2C-D). In addition, results of immunofluorescence revealed that there was no significant difference in the number of activated CD86+ CD11c+ dendritic cells among these groups before the intervention of anti-PD-L1 antibodies, whereas the number of these cells in each group increased after anti-PD-L1 antibody intervention (Supplementary Figure S3).
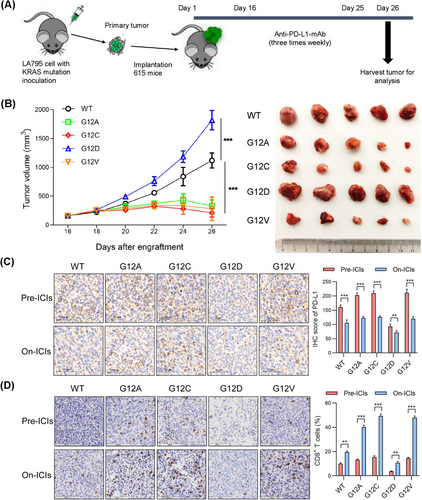
3.2 KRAS-G12D mutation contributed to an immune-suppressive TIME
Based upon 112 resected NSCLC samples with KRAS mutations (Supplementary Table S5), lower proportions of cells positive for PD-L1 protein expression and CD8+ T cell infiltration was observed in the G12D group compared with the non-G12D group (PD-L1: P = 0.003; CD8+ T cell: P = 0.043; Figure 3A-B). When the TIME was classified according to the PD-L1 status and the presence or absence of TILs [41, 42], significantly higher proportions of the TIME state with PD-L1-negative and no TILs was found in the G12D group (P = 0.009; Figure 3C), suggesting an immune-ignorance TIME in NSCLC samples with KRAS-G12D mutations.
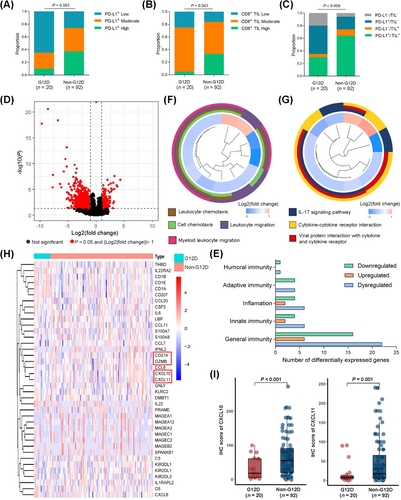
To explore the underlying mechanism of immunosuppressive TIME induced by the KRAS-G12D mutation, we analyzed the gene expression profile of 139 LUAD samples with KRAS mutations from the TCGA database. Among 1584 genes differentially expressed in LUAD specimens with or without G12D mutations, 39 immune genes were determined (Figure 3D-E). We then performed GO and KEGG enrichment analysis to determine the biological function and pathway correlating with the 39 differentially expressed genes. The results showed that these genes were enriched in the leukocyte chemotaxis pathway, leukocyte migration pathway, and cytokine-cytokine receptor interaction pathway (Figure 3F-G). Looking for differential genes related to adaptive immune response, significantly lower expression of genes related to effector T cell activation and killing tumor cells, including CD274, CXCL10, CXCL11 and Granzyme B (GZMB), were observed in the G12D group than in the non-G12D group (Figure 3H). CXCL10 and CXCL11, as chemokines, are involved in the recruitment of CD8+ T cells into tumor tissues, thereby resulting in the killing of tumor cells [43]. IHC analysis of the protein levels of CXCL10 and CXCL11 in the 112 resected NSCLC samples with KRAS mutations confirmed that the G12D group showed lower levels of CXCL10 and CXCL11 than the non-G12D group (CXCL10: P < 0.001; CXCL11: P = 0.001; Figure 3I).
3.3 KRAS-G12D mutation suppressed the infiltration of CD8+ T cells by regulating the HMGA2-CXCL10/CXCL11 axis
To explore the molecular mechanism by which the KRAS mutation regulates differential expression of CXCL10 and CXCL11 in the tumor microenvironment, we first planned to knock out the KRAS gene in Beas-2B cells which express wild-type KRAS gene (Figure 4A). However, only single-cell clones with heterozygous KRAS gene knockout survived (Figure 4B). Beas-2B cells with heterozygous KRAS gene knockout were then used to stably overexpress four common KRAS oncogene substitutions (Figure 4C). Of note, KEGG analysis of the biological processes and pathways enriched for the differential genes between the G12D-mutant and non-G12D-mutant cells suggested the involvement of cytokine-cytokine receptor interaction (Figure 4D). Next, we looked for the same significantly dysregulated genes between the LUAD samples and cell lines when comparing LUAD with KRAS-G12D mutations with other types of KRAS mutations, and identified 4 upregulated genes and 10 downregulated genes (Figure 4E-F). Then, a cross-correlogram was generated based on Pearson's correlation coefficient values among levels of PD-L1, CXCL10, CXCL11 and 14 significantly dysregulated genes in the LUAD samples with KRAS mutations. A markedly positive relationship between the HMGA2 level and that of PD-L1, CXCL10 and CXCL11 was found (Figure 4G). Considering that immune response is closely associated with the immune cell landscape, a TIMER algorithm was applied to analyze the relationship of HMGA2 level with the intra-tumoral immune cell composition. HMGA2 expression was positively related to the infiltration level of CD8+ T cells, neutrophils, myeloid dendritic cells and macrophages (Figure 4H). Furthermore, a positive correlation was confirmed through a combined analysis of the levels of HMGA2, PD-L1, CXCL10 and CXCL11 and the infiltration of CD8+ T cells in the IHC-detected group consisting of 112 resected NSCLC samples (Figure 4I). The IHC results of tumor tissues also demonstrated that the KRAS-G12D mutation induced low level of HMGA2 compared to the other mutations (P < 0.001; Figure 4J). Consistent with the IHC results obtained from tumor tissues, Western blotting and RT-qPCR in cells with four common KRAS oncogene substitutions further confirmed that the KRAS-G12D mutation led to low levels of HMGA2, CXCL10, CXCL11 and PD-L1 compared to other mutations (Figure 4K, Supplementary Figure S4A-D). The ELISA results also showed that KRAS-G12D mutation resulted in low secretion of CXCL10 and CXCL11 (Supplementary Figure S4E-F).

To verify the regulation mechanism between HMGA2, PD-L1, CXCL10 and CXCL11 levels, HMGA2-overexpressing Beas-2B-G12D, SK-LU-1, and A427 cell lines were analyzed. As speculated, HMGA2 expression, whether at the protein or messenger RNA (mRNA) level, upregulated levels of CXCL10 and CXCL11. HMGA2 expression also increased levels of secreted CXCL10 and CXCL11. Curiously, the level of PD-L1 was not regulated by HMGA2 expression (Figure 4L, Supplementary Figure S5).
3.4 KRAS-G12D mutation suppressed PD-L1 expression level via the P70S6K/PI3K/AKT axis
Previous studies have demonstrated that KRAS mutation stabilizes PD-L1 mRNA via the MEK-ERK signaling pathway [44]. However, a study reported that in addition to the MEK-ERK pathway, NSCLC cell lines with KRAS-G12D mutations had the activated PI3K-AKT pathway, whereas NSCLC cell lines with KRAS-G12V and KRAS-G12C mutations had the activated RalGDS-Ral pathway and the weakened activation of the PI3K-AKT pathway [27]. Hence, we speculated that different substitutions of the KRAS oncogene might differentially regulate PD-L1 level via the activation of different downstream signaling pathways.
Consistent with the previous study, KRAS mutation, regardless of the type of variation, promoted the phosphorylation of Erk1/2. However, compared to G12A, G12C and G12V point mutations, G12D point mutation activated the phosphorylation of AKT and reduced the phosphorylation of p70S6K (Figure 5A). These findings were also confirmed in LA795 cells with different substitutions of the KRAS oncogene (Supplementary Figure S6). Given that mechanistic target of rapamycin (mTOR) activation leads to feedback repression of the PI3K-AKT pathway through the effector p70S6K [45], KRAS-transfected Beas-2B cell lines were treated with the mTOR inhibitor (rapamycin, 0.5 μmol/L) for 24h, and the impact on the levels of PD-L1, HMGA2, CXCL10 and CXCL11, as well as the downstream signaling, were investigated. Rapamycin induced activation of the AKT pathway in Beas-2B cells expressing G12A, G12C and G12V point mutations. However, in Beas-2B cells expressing the G12D point mutation, the AKT pathway was constitutively activated, and rapamycin did not further increase the phosphorylation of AKT (Figure 5B). In addition, regardless of the type of KRAS variation, PD-L1 levels were reduced to a consistent level under the intervention of rapamycin (Figure 5B, Supplementary Figure S7A). However, after the intervention of rapamycin, the protein or mRNA levels of HMGA2, CXCL10 and CXCL11 as well as levels of secreted CXCL10 and CXCL11 in Beas-2B cells expressing the G12D point mutation remained lowest among the KRAS-transfected Beas-2B cell lines (Figure 5B, Supplementary Figure S7B-F).
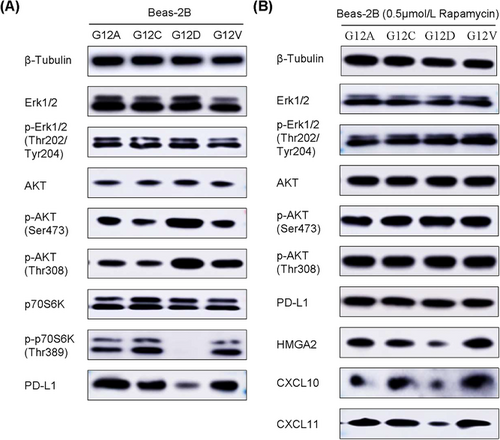
3.5 Paclitaxel-based chemotherapy recruited CD8+ T cells by upregulating CXCL10 and CXCL11 via HMGA2
A recent study showed that chemotherapy can induce tumor cells to secrete CXCL11 via the upregulation of HMGB1 level, thereby promoting the infiltration of CD8+ T cells in NSCLC [46]. Given that HMGA2 and HMGB1 are HMG family proteins, and paclitaxel is approved for the treatment of NSCLC, including LUAD and LUSC [47, 48], three cell lines harboring KRAS-G12D mutations (Beas-2B-G12D, SK-LU-1, A427) were treated with paclitaxel-based chemotherapy, and the effect on the levels of HMGA2, CXCL10 and CXCL11 were evaluated. The results revealed that paclitaxel increased the expression levels of HMGA2, CXCL10 and CXCL11 in a concentration-dependent manner (Figure 6A-C, Supplementary Figure S8). In addition, a patient-derived organoid (PDO) model with KRAS-G12D mutation was constructed and treated with paclitaxel (Supplementary Figure S9). Consistent with the results of the cells, the levels of HMGA2, CXCL10, and CXCL11 in PDO model with KRAS-G12D mutation were also upregulated in a concentration-dependent manner (Figure 6D, Supplementary Figure S10).
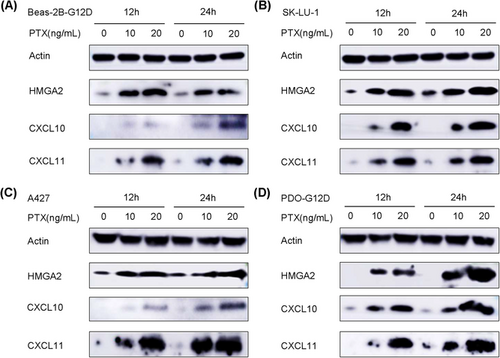
3.6 Chemo-immunotherapy as a treatment for KRAS-G12D-mutant NSCLC with primary resistance to ICI monotherapy
As chemotherapy could ameliorate the immunosuppressive TIME induced by the KRAS-G12D mutation by recruiting CD8+ T cells via upregulation of HMGA2, CXCL10 and CXCL11, we hypothesized that combined treatment with ICIs and chemotherapy would be a better option for NSCLC with KRAS-G12D mutation. A mouse model with KRAS-G12D mutation was established to receive anti-PD-L1 mAb or paclitaxel as a monotherapy or combined therapy (Figure 7A). Compared to the other groups, anti-PD-L1 mAb with paclitaxel remarkably suppressed tumor growth in the mice (P < 0.001; Figure 7B-C). IHC analysis showed that combined treatment significantly reduced PD-L1 protein levels, increased the protein levels of CXCL10 and CXCL11, and promoted the infiltration of CD8+ T cells (Figure 7D-G). Although the infiltration of CD8+ T cells and the expression of CXCL10 and CXCL11 increased after the intervention of paclitaxel, more abundant CD8+ T cells and more levels of CXCL10 and CXCL11 were observed in the combined treatment group (Figure 7E-G).
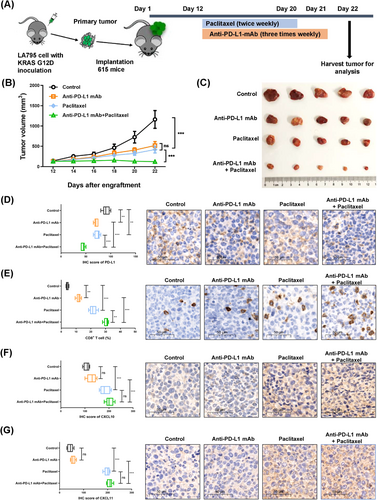
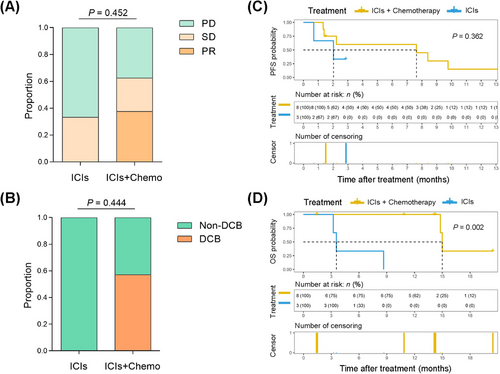
The above-mentioned mouse data paved the way for combination treatment with ICIs and chemotherapy as a rational, promising therapy to improve the immunosuppressive state and enhance the infiltration of CD8+ T cells in patients with KRAS-G12D-mutant NSCLC. Therefore, we retrospectively collected the clinical information of patients with NSCLC harboring KRAS-G12D mutation after ICI monotherapy or chemo-immunotherapy (Supplementary Table S6). In total, 11 patients were included to undertake treatment efficacy evaluations. Three (27.3%) patients received anti-PD-1 agent monotherapy, and 8 (72.7%) received chemo-immunotherapy. Three (27.3%) patients were identified with partial response, 3 (27.3%) patients with stable disease, and 5 (45.5%) with progressive disease (PD). Because the follow-up time of 2 (18.2%) patients was less than 6 months, 5 (45.5%) patients were determined to have DCB, and 4 (36.4%) to have non-DCB (PFS ≤ 6 months). Although there was no statistical difference, the proportion of PD and non-DCB was lower in the chemo-immunotherapy group compared to the ICI monotherapy group (Figure 8A-B). At the time of survival analysis, patients with NSCLC could obtain more benefit from chemo-immunotherapy with respect to PFS; however, there were no significant difference because of the small sample size (P = 0.362; Figure 8C). Of note, patients with KRAS-G12D-mutant NSCLC receiving chemo-immunotherapy had better overall survival (OS) than those with ICI monotherapy (P = 0.002; Figure 8D).
4 DISCUSSION
Although the successful application of small-molecule inhibitors targeting KRAS-G12C mutations in clinical trials has demonstrated that KRAS mutations are not “undruggable”, these drugs are not yet ready to enter clinical practice. Therefore, ICIs targeting the PD-1/PD-L1 axis, according to current guidelines, remains the optimal treatment for these patients [13, 49]. However, in contrast to other variants of KRAS mutations, our study found that the KRAS-G12D mutation drove immunosuppression and the primary resistance of immunotherapy against PD-1/PD-L1 by downregulating PD-L1 level and infiltration of CD8+ TILs in NSCLC.
Consistent with others, our study also proved that KRAS mutations were positively correlated with CD8+ TILs, PD-L1 level, and TMB, thereby resulting in an inflammatory TIME with adaptive immune resistance that is associated with a superior response to ICIs [17, 50]. However, NSCLC with KRAS mutation is a heterogeneous disease with different molecular characteristics, including distinct point variations and tumor-associated co-mutations [49]. Recent clinical studies have demonstrated that KRAS mutations do not perfectly guide immunotherapy in advanced NSCLC. First, emerging data indicate improved efficacy in patients with KRAS mutations and co-mutant TP53, and reduced efficacy in patients with KRAS mutations and co-mutant STK11/LKB1, KEAP1/NFE2L2 or SMARCA4 [51-55]. Second, although some previous studies have found that there are no significant differences in clinical benefit among patients with NSCLC harboring different variant statuses of the KRAS oncogene, the KEYNOTE042 study reported that the KRAS-G12C mutant subgroup receiving pembrolizumab monotherapy had a higher objective response rate, as well as longer PFS and OS, than other subgroups [56-60]. Interestingly, in an online clinical cohort of 74 patients with NSCLC, we found that KRAS-G12D mutations derived less benefit from ICI monotherapy. To confirm this finding, a co-culture system evaluating the tumoricidal activity of CD8+ T cells cocultured with KRAS-mutant tumor cells, as well as a LUAD mouse model with different KRAS oncogene substitutions, was established.
Previous studies have demonstrated that the predictive biomarkers of clinical benefit of NSCLC treated with ICIs include the TIME (PD-L1 level, CD8+ TILs, and other immune cells), genetic alterations (TMB, the loss and gain of activated mutations), and the host immune system [61, 62]. Consistent with the results of a recent publication [23], our study also found that patients with KRAS-G12D-mutant NSCLC tend to have lower TMB via in silico analysis of the clinical cohort and the TCGA database.
In addition, our study showed that the KRAS-G12D mutation was correlated with low level of PD-L1 and low infiltration of CD8+ TILs, inducing an immunosuppressive TIME. Several studies also analyzed the association between KRAS oncogene substitutions and PD-L1 level in tumor samples with NSCLC. Arbour et al. [57] stated that PD-L1 level was higher in patients with KRAS-G12C-mutant NSCLC compared to those with non-G12C mutations. Another study showed that KRAS-G12V mutation promoted PD-L1 level via the transcription of growth factor beta (TGF-beta)/epithelial-to-mesenchymal transition (EMT) pathway in NSCLC [30]. These conclusions do not contradict our finding that the KRAS-G12D mutation was negatively correlated with PD-L1 level. Although a previous study reported that KRAS mutation increased PD-L1 level via the MEK-ERK pathway [44], our study found that KRAS-G12D mutations not only activated the MEK-ERK signaling pathway but also suppressed PD-L1 level via the P70S6K/PI3K/AKT axis. These mechanisms explain why PD-L1 level of NSCLC with KRAS-G12D mutation was lower than other KRAS substitutions.
Of note, our study proposed that KRAS-G12D mutation suppressed the secretion of chemokines CXCL10/CXCL11 in TIME by downregulating the HMGA2 signaling, leading to a decrease in CD8+ TILs, which in turn built up a suppressive TIME that is primarily resistant to anti-PD-1/PD-L1 immunotherapy in NSCLC. In addition, some studies have reported the role of KRAS-G12D mutation, a common oncogene driver in these tumors, in the construction of TIME in pancreatic and colorectal cancers. Cheng et al. [63] found that KRAS-G12D mutation promoted the conversion of regulatory T cells by activating the MEK/ERK pathway, resulting in an immunosuppressive TIME in pancreatic cancer. Liao et al. [64] reported that KRAS-G12D mutation increased the infiltration of bone marrow-derived suppressor cells via the IRF2-CXCL3-CXCR2 axis, therapy leading to an immunosuppressive TIME that avoided T cell killing and caused resistance to immunotherapy in colorectal cancer.
According to current guidelines, chemo-immunotherapy is recommended as the first-line treatment for advanced NSCLC [65, 66]. However, from recent clinical data, ICIs combined with chemotherapy did not always increase the clinical benefit compared to ICIs alone in patients with KRAS-mutant NSCLC. Sun et al. [67] reported that among patients with KRAS mutations and PD-L1 level of 50% or greater, there was no difference in OS between those receiving ICI monotherapy and those receiving chemo-immunotherapy. Our previous study also demonstrated that anti-PD-L1 drugs combined with docetaxel did not extend the anti-tumor response in a mouse model with the KRAS-G12C mutation [17]. Interestingly, in the current research, ICI in combination with chemotherapy was shown to be more effective than ICI monotherapy in patients with KRAS-G12D-mutant NSCLC. We found that paclitaxel-based chemotherapy could recruit CD8+ T cells by upregulation of levels of CXCL10 and CXCL11 via the HMGA2 signaling, thereby meliorating the immunosuppressive TIME induced by the KRAS-G12D mutation. However, KRAS-mutant tumors with non-G12D mutations generally had high expression of PD-L1 and abundant TIL infiltration, suggesting a greater sensitivity to ICIs. Thus, when ICIs were combined with chemotherapy, the TIME was not effectively improved, the infiltration of CD8+ T cells was not increased, and additional clinical benefits were not observed in KRAS-mutant tumors with non-G12D mutations. Together, these results suggest a potential mechanism for the vulnerability of NSCLC with KRAS-G12D mutations to chemo-immunotherapy. However, the limitations of the present study should be recognized. For example, these findings were only verified by a preclinical mouse model and limited clinical data. Further validation should be performed in large prospective clinical trials in the future.
5 CONCLUSIONS
In conclusion, our study elucidated the molecular mechanism by which KRAS-G12D mutation drives immunosuppression and the primary resistance of immunotherapy against PD-1/PD-L1 in NSCLC. Moreover, we propose that a combination of ICIs and chemotherapy may be more effective in patients with KRAS-G12D-mutant NSCLC.
DECLARATIONS
ACKNOWLEDGEMENTS
Not applicable.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
Chengming Liu: project design, cell experiment, data curation, formal analysis, and writing-original draft preparation. Sufei Zheng, Zhanyu Wang, Sihui Wang, and Xinfeng Wang: mouse experiment and data analysis. Lu Yang, Haiyan Xu, and Zheng Cao: data curation and data analysis.
Xiaoli Feng, Qi Xue, and Yan Wang: project supervision and formal analysis. Jie He, Yan Wang, and Nan Sun: project administration, supervision, writing-reviewing and editing and funding acquisition. All authors read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The human study was approved by the Ethics Committee of Cancer Hospital and Institute, Chinese Academy of Medical Sciences (permit number: #20/242-2438). The tissue samples were obtained with written informed consent from each patient. The animal study was carried out in compliance with the guidance suggestion of Animal Care Committee of Cancer Hospital and Institute, Chinese Academy of Medical Sciences (permit number: #NCC2020A163).
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The datasets used and/or analyzed in the present study can be obtained from the corresponding authors as reasonably required.




