Proteomics profiling of colorectal cancer progression identifies PLOD2 as a potential therapeutic target
Yingkuan Shao and Kailun Xu contributed equally to this work.
Abbreviations
-
- AC
-
- adenocarcinoma not otherwise specified
-
- CRC
-
- colorectal cancer
-
- DIA-MS
-
- data-independent acquisition mass spectrometry
-
- ECM
-
- extracellular matrix
-
- FFPE
-
- formalin-fixed paraffin-embedded
-
- IHC
-
- immunohistochemistry
-
- IPA
-
- ingenuity pathway analysis
-
- KO
-
- knockout
-
- MC
-
- mucinous carcinoma
-
- NC
-
- normal control
-
- PCT
-
- pressure cycling technology
-
- PDXs
-
- patient-derived xenografts
-
- PLOD2
-
- Procollagen-Lysine, 2-Oxoglutarate 5-Dioxygenase 2
-
- PRM
-
- parallel reaction monitoring
-
- TMA
-
- tissue microarray
-
- WB
-
- western blotting
Colorectal cancer (CRC) is the second leading cause of cancer deaths in developed countries [1]. The malignant transformation from small clumps to cancer takes about 10 years [2]. This study aimed to characterize proteomic dynamics associated with CRC development and progression, and identify novel therapeutic targets for intercepting the underlying oncogenic processes. We have optimized pressure cycling technology (PCT) coupled with data-independent acquisition mass spectrometry (DIA-MS) for robust and reproducible proteomic analysis of biopsy-level formalin-fixed paraffin-embedded (FFPE) tissues [3].
In this study, we profiled the proteomic tissue landscape of CRC evolving from normal colon to hyperplastic polyps, adenomas, adenocarcinoma not otherwise specified (AC) or mucinous adenocarcinoma (MC). We identified 69,949 peptides, 6,359 protein groups, and 4,830 unique proteins (Supplementary Table S1) based on our previously established spectral library for DIA analysis [4] from 170 FFPE tissue samples (85 patients, each with 2 biological replicates) (Figure 1A). Pearson's correlation coefficient between biological replicates was 0.813, and 0.953 between technical replicates.
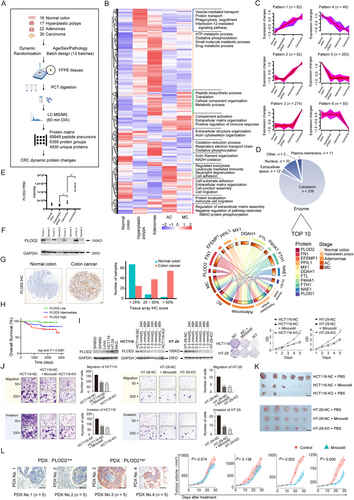
We identified 928 differentially expressed proteins by comparing protein expression in samples from different CRC clinical stages to normal colon tissue samples (Figure 1B). Pairwise comparisons between polyps and normal colon, adenomas and polyps, carcinoma and adenomas, as well as MC and AC revealed distinct proteomic changes associated with each transformation towards malignancy (Supplementary Figure S1A). Canonical pathways analysis revealed that the dysregulated proteins were mostly related to oxidative phosphorylation. Interestingly, oxidative phosphorylation was enhanced in precancerous tissues (hyperplastic polyps and adenomas) but suppressed in CRC tissues, suggesting metabolic adaptations of tumor cells in the evolving microenvironment (Supplementary Figure S1B). Analysis of diseases and biological functions of differential proteins in benign lesions showed proteomic perturbations associated with oncogenic pathways. For example, COPE, COPA, and COPZ1 are proteins encoded by coatomer protein complex genes which are essential proteins for tumorigenesis in CRC. PSMC3, PSMD13, PSMA7 and PSMD8 are all proteasomal proteins whose expressions began to rise in polyps and peaked in adenomas (Supplementary Figure S1C).
Six biologically significant protein expression patterns associated with CRC development were selected by unsupervised cluster analysis. Patterns 1, 2 and 3 were formed by upregulated proteins while patterns 4, 5 and 6 were formed by downregulated proteins (Figure 1C). Gene Ontology analysis for enrichment of biological processes in the six clusters (Supplementary Figure S2A) identified extracellular matrix (ECM) enrichment in pattern 3. We then checked for all ECM-related proteins in the “matrisome”, which has been defined as the combination of core ECM proteins (glycoproteins, collagens, and proteoglycans) and ECM-associated proteins (ECM-affiliated proteins, ECM regulators, and secreted factors) [5]. Among the six protein expression patterns, we observed enrichment of ECM regulators in the upregulated patterns. Proteins within each pattern formed protein-protein interaction networks using Cytoscape with the GeneMania plugin (Supplementary Figure S2B).
We then narrowed our focus on proteins that were consistently upregulated along the stages of tumor progression. Among plasma membrane, nucleus, cytoplasm, extracellular space and other locations, our data showed that the cytoplasm and extracellular proteins stood out as the locations with the highest expression of dysregulated proteins (Supplementary Figure S2C). CRC progression was associated with substantially increased expression of multiple enzymes (Supplementary Figure S3A). Of note, Procollagen-Lysine, 2-Oxoglutarate 5-Dioxygenase 2 (PLOD2) was the most up-regulated protein (Figure 1D and Supplementary Figure S3B). We further performed a pairwise comparison between pre-cancerous and cancerous samples with benign samples, and PLOD2 consistently outstood as the top hit (Supplementary Figure S3C).
Next, we randomly selected eight samples of each tissue type (normal colon, hyperplastic polyps, adenomas, AC, and MC) for targeted measurement of PLOD2 using parallel reaction monitoring (PRM). The PRM data from the 40 samples confirmed the elevation of PLOD2 (Figure 1E). As further verification, western blot (WB) analysis was performed on four new CRC patients and observed higher PLOD2 expression in CRC tissues than in matched para-tumoral normal colon (Figure 1F). We also assessed PLOD2 expression by immunohistochemistry staining (IHC) of tissue microarrays (TMAs) containing 118 CRC (8th AJCC TNM Stage II) and 79 para-tumoral normal colon tissues (Supplementary Table S2). The IHC staining of PLOD2 in para-tumoral normal colon tissues showed that 68 samples had < 25% positive colon cells, eight samples had 25%-50%, and only three samples had > 50%. In contrast, CRC tissues showed significantly higher PLOD2 expression than para-tumoral normal colon (Figure 1G). Remarkably, we found that higher PLOD2 expression was associated with poorer overall survival of CRC patients (Figure 1H, Supplementary Table S2).
Next, we measured the PLOD2 expression in six CRC cell lines and chose the two with the highest PLOD2 expression, namely HCT116 and HT-29, for generating PLOD2-knockout (KO) congener lines using CRISPR-Cas9. Each congenic pair of cell lines was treated with increasing concentrations of minoxidil, a lysyl hydroxylase inhibitor of PLOD2. Minoxidil inhibited PLOD2 expression in both wild-type cell lines in a time- and dose-dependent manner, while clonogenicity and cell proliferation were dramatically suppressed when PLOD2 was inhibited by minoxidil or knocked-out (Figure 1I). In addition, both minoxidil treatment and PLOD2-KO suppressed CRC cell migration and invasion (Figure 1J).
Extending our findings to in vivo model, we injected HCT116 and HT-29 cell lines subcutaneously into nude mice. We tested the effects of placebo versus minoxidil treatment in vivo on tumors generated by wild-type HCT116 and HT-29 cells, and also compared the growth of tumors generated by PLOD2-KO HCT116 and HT-29 cells with PLOD2-high tumors. Our data showed that both minoxidil and CRISPR-Cas9-mediated PLOD2 suppression led to a significant decrease in tumor volume (Figure 1K). A second in vivo model was patient-derived xenografts (PDX) tumors from four CRC patients. Patient tumors with high PLOD2 levels (PLOD2 positive tumor cells were > 80%) were sensitive to minoxidil inhibition while PLOD2-negative tumors were resistant (Figure 1L), highlighting the potential clinical application of targeted therapy against PLOD2.
To gain mechanistic insight on how PLOD2 inhibition suppresses CRC tumors, we compared the transcriptome and proteome of HCT116-KO and the HCT116-normal control (NC) cell lines using RNA sequencing and DIA-MS, respectively (Supplementary Table S3). We identified 1236 up- and 955 down-regulated transcripts, and 227 up- and 127 down-regulated proteins (Supplementary Figure S4A). The data indicated that PLOD2 contributed to tumor growth, resistance to cell necrosis, and was closely related to the development of colorectal cancer (Supplementary Figure S4B). PLOD2 was also involved in protein synthesis, metabolism, and mRNA translation (Supplementary Figure S4C). Selected protein networks prioritized by these analyses are shown in Supplementary Figure S4D and E. An overview of the patients' basic pathological characteristics is shown in Supplementary Table S4.
Compared to previous studies [6, 7], our study systematically tracked a plethora of protein changes in CRC tissues as the disease progressed through increasing degrees of malignancy. Therapeutic interventions directed at cancer-derived ECM and their regulatory factors may be clinically effective [8]. PLOD family proteins catalyze post-translational modifications of collagen by converting lysine to hydroxylysine, which promotes stable interactions and deposition of collagen [9]. PLOD2 could be induced in L1CAM-overexpressing CRC cell lines and promoted L1CAM-mediated CRC progression by inducing ezrin signaling and the SMAD2/3 pathway [10]. Our data collectively constitute plausible evidence for suggesting further research on PLOD2 as a promising therapeutic target in CRC tumors in the emerging practice of precision oncology.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Written informed consent was obtained from all participants based on the guidelines of the Declaration of Helsinki. Human tissue samples were collected with the approval of the Institutional Ethics Committee of the Second Affiliated Hospital of Zhejiang University, School of Medicine (Zhejiang, P. R. China, No.2020-322). Animal studies and formalin-fixed paraffin-embedded samples collections were approved by the Institutional Ethics Committee of the Second Affiliated Hospital Zhejiang University School of Medicine (SYXK2018-0012).
CONSENT FOR PUBLICATION
All authors read and approved the final manuscript for publication.
AVAILABILITY OF DATA AND MATERIAL
The MS proteomics data are available on the iProX database with the project ID: IPX0001414000 and the subproject ID: IPX000141400. The raw sequence data have been uploaded to SRA with an ID: PRJNA598559.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This work was supported by the National Key Research and Development Program of China (Grant No. 2017YFC0908200), National Natural Science Foundation of China (Grant No. 81972270, 81972492, 32027801, 21904107), the Zhejiang Provincial Science Foundation for Distinguished Young Scholars (Grant No. LR19C050001), Hangzhou Agriculture and Society Advancement Program (Grant No. 20190101A04) and 2019 Zhejiang University Academic Award for Outstanding Doctoral Candidates to YK.S (Grant No. 2019071).
AUTHORS’ CONTRIBUTIONS
YKS, KLX, XZ, BTZ, XLZ, LW, YTS, DL performed experiments and data interpretation. TC, JW, SJY, LFS, XMX, LNQ, JNC, WXH, XYW, XPX, JFL, LRC, JMS, SZ provided key biological samples and materials. YKS, KLX, LW, DL, SZD, HHG, GR, YTS, WL, XC, TSZ, YZ, ZYH, TNG performed data analysis. YKS, KLX, XZ, BTZ, YTS, SZ, JMS, TNG designed the study, interpreted the data, and wrote the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
We particularly acknowledge the Biobank of the Second Affiliated Hospital of Zhejiang University School of Medicine. We thank Westlake University Supercomputer Center for assistance in data storage and computation and the Biomedical Core facility for mass spectrometry analysis. We thank Prof. Yongzhan Nie (Air Force Medical University) for his guidance in writing. We thank Dr. Oi Lian Kon for editing the manuscript.




