The scaffold protein menin is essential for activating the MYC locus and MYC-mediated androgen receptor transcription in androgen receptor-dependent prostate cancer cells
Abbreviations
-
- AR
-
- Androgen Receptor
-
- ARlow/CD44+
-
- with low AR expression and positive CD44 expression
-
- ASH2L
-
- ASH2 like
-
- ChIP
-
- Chromatin Immunoprecipitation
-
- Chr 1
-
- Chromosome 1
-
- EZH2
-
- Enhancer of zeste homolog 2
-
- KMT2A
-
- Lysine-Specific Methyltransferase 2A
-
- KMT2B
-
- Lysine-Specific Methyltransferase 2B
-
- LncRNA
-
- Long non-coding RNAs
-
- MEN1
-
- Multiple endocrine neoplasia type 1
-
- mCRPC
-
- metastatic castration-resistant PCa
-
- PCa
-
- prostate cancer
-
- PCAT1
-
- Prostate Cancer Associated Transcript 1
-
- siRNA
-
- Small interfering RNA
Dear Editor,
The proto-oncogene MYC has a well-documented role in driving androgen receptor (AR) oncogenic functions in prostate cancer (PCa cells) [1] and was recently reported as a key activator in the regulation of AR transcription [2]. Dissecting the mechanisms underlying MYC-mediated AR regulation may be crucial for better understanding PCa development and improving its management. We recently demonstrated that menin, encoded by the MEN1 gene, plays an oncosuppressive role in the generation of ARlow/CD44+ microinvasive prostate adenocarcinoma in mice but exerts oncogenic effects specifically in AR-dependent human PCa cells, both likely by modulating AR transcription and AR targets [3]. We herein wondered whether this oncogenic function of menin would occur through its interaction with other factors known to play a crucial role for AR transcription in PCa, in particular, MYC.
We initially examined the expression of MYC upon MEN1 knockdown (KD) in PCa cells. We found that MEN1 silencing led to a decrease in MYC expression at transcriptional (Fig. 1a, Supplementary Fig. S1A) and protein levels (Fig. 1b, Supplementary Fig. S1B-D) in AR-dependent (VCaP, LNCaP, and CWR22Rv1), but not AR-independent (PC3 and DU145), PCa cells. We also examined MYC expression in mouse prostate and found no altered MYC expression in menin-deficient, compared to menin-expressing, luminal cells (Fig. S1E). Therefore, we hereafter focused on AR-dependent PCa cells. We detected the interaction between menin and MYC in AR-dependent PCa cells (Supplementary Fig. S2A-B), similar to HeLa cells [4], and found that MYC-KD triggered reduced cell proliferation in these cells (Supplementary Fig. S2C-D). Recently, several distal regulatory sequences in the MYC locus have been identified, including 2 enhancers situated 67 kb upstream [5] and 20 kb downstream [6] of the transcription start site, respectively named MYC enhancer 1 and 2 for the current study. Importantly, ChIP analyses demonstrated that menin bounded not only to the MYC promoter but also to these two enhancers (Fig. 1C, Supplementary Fig. S3A). Moreover, menin was necessary for major components of the MLL complex (KMT2A, KMT2B, ASH2L), to which menin belongs [7], to bind to and activate the MYC promoter and enhancer 2 likely via H3K4me3 marks (Fig. 1D-E, Supplementary Fig. S3B-E). MEN1 silencing resulted in decreased expression of known MYC target genes, enhanced cell cycling alterations (Supplementary Fig. S4A-C), and decreased expression of AR variants (Supplementary Fig. S4D-E), reminiscent of previous findings in other organs [8]. Notably, the expression of menin and MYC, their interaction, and the binding of menin to the MYC promoter and enhancer 2, were augmented upon stimulation with the androgen, dihydrotestosterone (Fig. 1F-H, Supplementary Fig. S5). Hence, these data revealed that menin is a crucial factor positively regulating MYC transcription specifically in AR-dependent PCa cells.
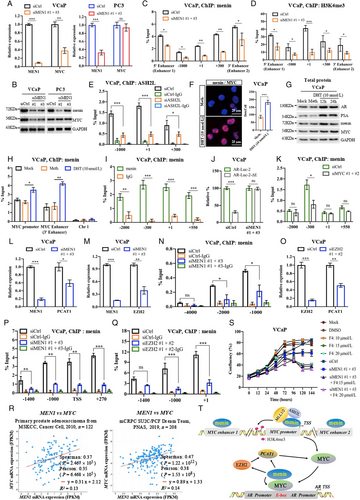
Menin exerts a regulatory role on the MYC locus and is essential for AR transcription mediated by MYC in AR-dependent PCa cells. MYC mRNA and protein levels were analyzed by RT-qPCR (A), Western blotting analysis (B) in siCtrl or siMEN1 #1 and siMEN1 #3-co-transfected VCaP and PC3 cells. C-K The experiments were conducted exclusively in AR-dependent VCaP cells. ChIP-qPCR assessing the binding of menin to (C) and H3K4me3 marks (D) on the MYC promoter and enhancers in siMEN1 #1 and siMEN1 #3-co-transfected cells. (E) ChIP-qPCR assessing the binding of menin to the MYC promoter in siCtrl or siASH2L-treated cells. (F) Proximity ligation assay (PLA) showing menin and MYC interaction upon DHT treatment for 24 h. Scale bar, 25 μm. Quantification of PLA signals per cell is shown on the right. (G) Western blotting analysis of AR, menin, and MYC levels upon treatment with DHT for 12 h and 24 h. (H) ChIP-qPCR showing the binding of menin to the MYC promoter and enhancer 2 following treatment with DHT for 24 h. Chr 1 was used as the negative control. (I) ChIP-qPCR of the binding of menin to the AR promoter. (J) Relative luciferase activity after transfection of siCtrl- or siMEN1 #1 and siMEN1 #3-co-transfected cells with AR-Luc-2 or AR-Luc-2-ΔE. (K) ChIP assessing menin occupancy on the AR promoter in siCtrl or siMYC #1 and #2-co-transfected VCaP cells as indicated. (L) RT-qPCR detecting PCAT1 expression in siCtrl or siMEN1 #1 and siMEN1 #3-transfected cells. (M) RT-qPCR showing EZH2 expression in siCtrl or siMEN1 #1 and siMEN1 #3-co-transfected cells. (N) ChIP evaluating menin occupancy on the EZH2 promoter. (O) RT-qPCR detecting PCAT1 expression in siCtrl or siEZH2 #1 and #2-co-transfected cells. (P) ChIP showing menin occupancy on the PCAT1 promoter. (Q) ChIP showing the effect of siEZH2 #1 and 2 on the levels of menin recruitment to the PCAT1 promoter. (R) Data mining analysis of the correlation between MEN1 and MYC mRNA expression in primary PCa (left panel) and mCRPC (right panel). (S) Combined use of MYC inhibitor (10058-F4) and siMEN1 #1 and 3 showing effects on cell proliferation. (T) Schematic summary of uncovered functions of menin in AR-dependent PCa cells. For all the RT-qPCR and ChIP-PCR analyses, shown is the mean ± s.d. (n = 3). t-test, ns, non-significant, * P < 0.05, ** P < 0.01, *** P < 0.001.
Abbreviations: AR, androgen receptor; ChIP, chromatin immunoprecipitation; Chr 1, Chromosome 1; Ctrl, Control; DAPI, 4′,6-diamidino-2-phenylindole; EZH2, Enhancer of zeste homolog 2; HPRT, Hypoxanthine-guanine phosphoribosyltransferase; MEN1, Multiple endocrine neoplasia type 1; PCa, prostate cancer; PCAT1, Prostate Cancer Associated Transcript 1; PCR, Polymerase chain reaction; RT-qPCR, quantitative real-time PCR; si, interfering RNA.
Next, we investigated whether menin was involved in MYC-mediated activation of AR transcription [2]. ChIP analysis and Luciferase reporter assays revealed that menin bounded to and positively regulated several sites on the AR promoter, whereas MYC had a greater binding affinity and regulatory activity for an E-box site located -300 kb of the AR promoter (Fig. 1I, Supplementary Fig. S6). Menin is also bounded to the KLK3 enhancer (Supplementary Fig. S6D). Importantly, deletion of the E-box on the AR promoter significantly reduced the transactivation activity of MYC and that of menin in Luciferase reporter assays whereas this latter activity remained unaffected on other parts of the AR promoter (Fig. 1J, Supplementary Fig. S6D-G, Supplementary Fig. S7A-C). Remarkably, we found that MYC silencing resulted in less menin binding to the E-box on the AR promoter (Fig. 1K, Supplementary Fig. S7D-E). Our data indicate, for the first time, that menin exerts a regulatory role on the MYC-mediated AR transcription.
In parallel, we assessed whether menin is involved in the regulation of LncRNA PCAT1, situated upstream of the MYC locus and known to play an oncogenic role through MYC in PCa [9]. Markedly, MEN1 silencing led to reduced PCAT1 expression in AR-dependent PCa cell lines (Fig. 1L, Supplementary Fig. S8A). Since EZH2 has been reported to directly regulate PCAT1 expression and that the interaction between EZH2 and menin has been described in different contexts [10], we investigated whether menin controls PCAT1 transcription through EZH2 in PCa cells. Here, menin was crucial for the expression of EZH2 (Fig. 1M, Supplementary Fig. S8B), via its physical interaction with EZH2 (Supplementary Fig. S8C-D) and its binding to the EZH2 promoter (Fig. 1N-O, Supplementary Fig. S8E-F). Finally, ChIP analysis showed that menin bound to the same binding sites as EZH2 on the regulatory sequences of PCAT1 (Fig. 1P, Supplementary Fig. S8G), the binding being weakened upon EZH2-KD (Fig. 1Q, Supplementary Fig. S8H). Taken together, our work unveiled that menin is critically involved in the regulation of MYC-related PCAT1, through its interaction with and regulation of EZH2.
Finally, through data mining, significant positive correlations were observed between MEN1 and MYC mRNA expression (Fig. 1R) and between MEN1 and EZH2 mRNA expression in both primary PCa and mCRPC (Supplementary Fig. S9). Lastly, the combined use of siMEN1 and the MYC/Max dimerization inhibitor 10058-F4 had a synergistic effect on reducing the proliferation of VCaP, LNCaP, and CWR22Rv1 cells (Fig. 1S, Supplementary Fig. S10).
Collectively, we uncovered a critical crosstalk between menin, AR, and MYC, involving PCAT1 regulation via EZH2, specifically in AR-dependent PCa cells (Fig. 1T), providing novel clues as to the mechanisms underlying the regulation of AR transcription.
DECLARATIONS
FUNDING
This study was supported by Epigenetics & Cancer Program (ASC14092CSA), the Ligue Inter-régionale contre le Cancer (R19040CC), Institut National du Cancer (PRTK2017) and the Association: «Le Cancer du sein, parlons-en». R.T was the recipients of a PhD-fellowship from Ministère de l'Enseignement Supérieur et de la Recherche, France. Y.L was the recipient of a PhD-fellowship from the China Scholarship Council. R.A.Z was the recipient of a PhD-fellowship from the Association “G04MEDIA S.A.R.L”, Lebanon.
ACKNOWLEDGMENTS
We are grateful to Brigitte Manship for her assistance in editing and proofreading the manuscript, Zhi Chong Wu for his scientific and technical input, and Silvère Baron for his scientific discussion and advice.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All animal experiments were performed following the animal care guidelines of the European Union and were validated by the local Animal Ethics Evaluation Committee (CECCAPP: C2EA-15 agreed by the French Ministry of High School and Research, « Autorisation de projet » CLB-2012-053).
AUTHORS’ CONTRIBUTIONS
Y.K.L conducted the experiments, analyzed and interpreted the data, prepared figures, and manuscript; V.V.G, T.R, and R.A.Z provided technical and material support and participated in data interpretation; P.B participated in study design and provided critical revision of the manuscript; M.L.R and C.X.Z conceived and supervised the study and manuscript preparation, and obtained funding.
CONSENT FOR PUBLICATION
All of the authors approved the contents and the data in this manuscript and agreed to its submission for publication.
CONFLICT OF INTERESTS
The authors declare that they have no conflict of interest.
AVAILABILITY OF DATA AND MATERIAL
All data generated or analyzed during this study are included in this article.




