An integrated genetic-epigenetic analysis shed light on the mechanisms linking coronavirus disease 2019 (COVID-19) and cancer
Abbreviations
-
- ACE2
-
- Angiotensin-converting enzyme 2
-
- BLCA
-
- Bladder carcinoma
-
- BRCA
-
- Breast invasive carcinoma
-
- CESC
-
- Cervical squamous cell carcinoma and endocervical adenocarcinoma
-
- COAD
-
- Colon adenocarcinoma
-
- COVID-19
-
- Coronavirus disease 2019
-
- COVID-19-SNP
-
- COVID-19 susceptibility SNP
-
- CRC
-
- Colorectal cancer
-
- CTSL
-
- Cathepsin L
-
- ESCA
-
- Esophageal carcinoma
-
- FDR
-
- False discovery rate
-
- FYCO1
-
- FYVE and coiled-coil domain autophagy adaptor 1
-
- GWAS
-
- Genome-wide association study
-
- HGI
-
- COVID-19 host genetics initiative
-
- HNSC
-
- Head and neck squamous cell carcinoma
-
- KIRC
-
- Kidney renal clear cell carcinoma
-
- KIRP
-
- Kidney renal papillary cell carcinoma
-
- LAML
-
- Acute myeloid leukemia
-
- LGG
-
- Brain lower-grade glioma
-
- LIHC
-
- Liver hepatocellular carcinoma
-
- LUAD
-
- Lung adenocarcinoma
-
- LUSC
-
- Lung squamous cell carcinoma
-
- NS3BP
-
- NS3 binding protein
-
- PAAD
-
- Pancreatic adenocarcinoma
-
- PCPG
-
- Pheochromocytoma and paraganglioma
-
- PRAD
-
- Prostate adenocarcinoma
-
- RCC
-
- Renal cell carcinoma
-
- RPLP2
-
- Ribosomal protein lateral stalk subunit P2
-
- SARC
-
- Sarcoma
-
- SARS-CoV-2
-
- Severe acute respiratory syndrome coronavirus 2
-
- SKCM
-
- Skin cutaneous melanoma
-
- SNP
-
- Single nucleotide polymorphism
-
- STAD
-
- Stomach adenocarcinoma
-
- TALDO1
-
- Transaldolase 1
-
- TCGA
-
- The cancer genome atlas
-
- TGCT
-
- Testicular germ cell tumors
-
- THCA
-
- Thyroid carcinoma
-
- THYM
-
- Thymoma
-
- TMPRSS2
-
- Transmembrane serine protease 2
-
- UCEC
-
- Uterine corpus endometrial carcinoma
The Coronavirus disease 2019 (COVID-19) is an emerging disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection [1, 2]. It has spread rapidly across the world and caused a global health emergency. As of 10 February 2021, COVID-19 has caused 2,333,446 deaths worldwide. The majority of COVID-19 deaths had comorbidities including cancer [1, 2]. Notably, a recent study suggested that cancer was the only independent risk factor for death of COVID-19 patients among common comorbidities [3]. Moreover, cancer patients are more susceptible to SARS-CoV-2 infection than noncancer individuals due to their specific immunosuppressive state [3]. Despite the links between COVID-19 and cancer, the genetic and epigenetic mechanisms underlying these links remain unclear. Thus far, only genetic and epigenetic variants within three COVID-19 receptors, angiotensin-converting enzyme 2 (ACE2), transmembrane serine protease 2 (TMPRSS2), and cathepsin L (CTSL), have been investigated in cancers [4]. However, the majority of COVID-19-related genetic and epigenetic variants still remain uninvestigated. Here, we performed a systematic genetic-epigenetic analysis to explore the mechanisms underlying the links between COVID-19 and cancer.
To decipher the potential mechanisms, we first identified COVID-19-susceptible single nucleotide polymorphisms (COVID-19-SNPs) and then assessed genetic-epigenetic links between COVID-19 and cancer using COVID-19-SNPs and (1) CpG methylation or (2) gene transcription/translation data (Figure 1a). To identify COVID-19-SNPs, we selected SNPs from two genome-wide association studies (GWAS) of COVID-19 and then filtered SNPs having a P value threshold of 1.0 × 10–5. There were 265 and 184 COVID-19-SNPs identified from the two GWAS studies, respectively (Figure 1b). Since the two GWAS studies were performed in large sample sizes (both n > 3,000), we merged the two sets of SNPs and ultimately identified 426 COVID-19-SNPs (Supplementary Table S1). The detailed data analysis process for this study is described in Supplementary Materials and Methods.
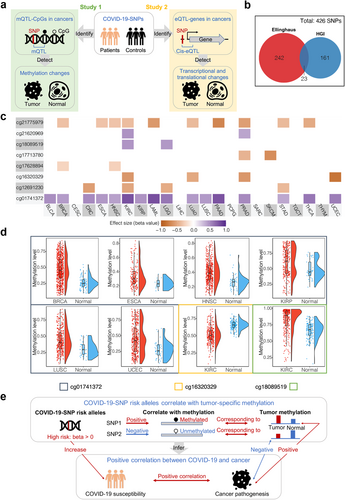
To investigate the molecular dependencies between COVID-19 and cancer using COVID-19-SNPs and methylation data, we first investigated interplays between COVID-19-SNPs and CpGs in various cancers. Across 23 cancer types in the Cancer Genome Atlas (TCGA), eight CpGs associated with COVID-19-SNPs were identified as methylation quantitative trait loci corresponding CpGs (mQTL-CpGs) in 17 types of cancer with a threshold of false discovery rate (FDR) < 0.05 and |r| ≥ 0.3 (Figure 1c, Supplementary Table S2). Amongst them, three mQTL-CpGs (cg21620969, cg18089519, and cg01741372) were positively correlated with COVID-19-SNP risk alleles in multiple cancers, whereas the remaining mQTL-CpGs were negatively correlated. Subsequently, we identified methylation changes in mQTL-CpGs between tumor and normal tissues. To obtain mQTL-CpGs that are more likely to capture marked cancer pathogenesis and to be devoid of mQTL-CpGs that show environmentally induced variability, we selected mQTL-CpGs with significant methylation changes greater than 10% in tumors versus normal tissues, since environmentally induced methylation changes rarely surpass 10%. Based on this approach, we yielded three mQTL-CpGs: cg01741372 in breast invasive carcinoma (BRCA), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney renal papillary cell carcinoma (KIRP), lung squamous cell carcinoma (LUSC), and uterine corpus endometrial carcinoma (UCEC); cg16320329 in kidney renal clear cell carcinoma (KIRC); and cg18089519 in KIRC (Figure 1d, Supplementary Table S3). cg01741372 and cg18089519 had higher methylation levels while cg16320329 had lower methylation levels in tumors versus normal tissues. Collectively, based on the directions of correlations demonstrated above, the positive correlation between COVID-19-SNP risk alleles and multiple cancer pathogenesis might be mediated by CpG methylation (Figure 1e).
Interestingly, all three identified mQTL-CpGs showed significant methylation changes between tumor and normal tissues in the major subtypes (i.e. KIRC and KIRP) of renal cell carcinoma (RCC), which is the most common (85%) malignant kidney tumor. This might suggest a potential mechanism underlying the links of COVID-19 and RCC observed in a previous epidemiological study [5]. The three mQTL-CpGs were separately located in gene body regions of NS3 binding protein (NS3BP), FYVE and coiled-coil domain autophagy adaptor 1 (FYCO1), and transaldolase 1 (TALDO1) (Supplementary Tables S3 and S4). NS3BP and TALDO1 are associated with chronic kidney disease, which is a risk factor for both RCC incidence and COVID-19 mortality [6]. Additionally, TALDO1 encodes a rate-limiting enzyme involved in the pentose phosphate pathway and shows increased levels in SARS-CoV-2-infected cells [7]. The pentose phosphate pathway contributes to various biological processes, including the ribonucleotide supplying for SARS-CoV-2 replication and carcinogenesis [7]. FYCO1 encodes a Rab7 adapter protein involved in autophagy, which is linked to SARS-CoV-2 infection and tumor development [8]. Specifically, autophagy substantially controls immune responses by regulating the production of cytokines and the functions of immune cells [8]. The immune system demonstrates complex interactions with kidney tumors, and its dysfunction can make it harder to fight the coronavirus infection [1, 2]. Collectively, methylation changes in these genes might be associated with specific biological functions and suggested as a mediator linking COVID-19 and RCC.
To explore the molecular dependencies between COVID-19 and cancer using COVID-19-SNPs and gene transcription/translation data, we initially identified genes correlated with COVID-19-SNPs in transcription levels (Figure 1a). Through a database analysis, 13 genes were identified (FDR < 0.05; Supplementary Figure S1a, Supplementary Table S5). Next, we tested their differential levels at both transcription and translation between tumor and normal tissues to ensure transcriptional changes leading to translational changes. Amongst the 13 candidate genes, ribosomal protein lateral stalk subunit P2 (RPLP2) showed significantly higher expression levels in tumor tissues of KIRC and lung adenocarcinoma (LUAD) (FDR < 0.05; Supplementary Figure S1b and c, Supplementary Table S6). Given that RPLP2 transcription levels in KIRC and LUAD could be elevated by the COVID-19-SNP risk allele, the positive correlation between COVID-19-SNP risk alleles and the pathogenesis of KIRC and LUAD could also likely be mediated by gene transcription/translation besides CpG methylation (Supplementary Figure S1d).
LUAD is the most frequent cancer type among these COVID-19 patients with malignancies. Furthermore, lung cancer patients had a higher incidence of COVID-19 and a greater risk of mortality from COVID-19 compared to the general population [1]. Here we demonstrated the elevated transcription and translation levels of RPLP2 in LUADs, which is consistent with a previous study (Supplementary Table S4) [9]. RPLP2 is related to the poor overall survival of LUAD patients [9]. Moreover, RPLP2 encodes a ribosomal protein involved in ribosome biogenesis, which can influence innate immune responses to virus infection and shows a hyperactive state in COVID-19 patients [10]. Collectively, it could be reasonable to suggest a mediation role of RPLP2 expression in linking COVID-19 and LUAD.
We conclude that methylation or transcription/translation levels of four genes served as the molecular dependencies underlying the positive links between COVID-19 and multiple cancers, especially RCC and LUAD. These findings were based on association studies on a large dataset from TCGA, and further biological experiments are still needed to confirm the proposed mechanisms.
ACKNOWLEDGMENTS
We would like to sincerely thank Professor Michael S. Kobor for all the knowledge and moral support.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST STATEMENT
The authors declare that there are no conflicts of interest.
AUTHOR CONTRIBUTIONS
ZD and HY conceived and designed the experiments. ZD and HY performed the experiments and analyzed the data. HY and ZD wrote the paper. All authors have read and approved the final manuscript.
AVAILABILITY OF DATA AND MATERIAL
All data generated or analyzed during this study were acquired from these databases: The COVID-19 HGI (https://www.covid19hg.org/results/), COVID-19 GWAS Results (https://grasp.nhlbi.nih.gov/Covid19GWASResults.aspx), TCGA (https://tcga-data.nci.nih.gov), Pancan-meQTL (http://bioinfo.life.hust.edu.cn/Pancan-meQTL/), UALCAN (http://ualcan.path.uab.edu/) and PancanQTL (http://bioinfo.life.hust.edu.cn/PancanQTL/).




