Mex3a promotes oncogenesis through the RAP1/MAPK signaling pathway in colorectal cancer and is inhibited by hsa-miR-6887-3p
Abstract
Background
Although Mex3 RNA-binding family member A (Mex3a) has demonstrated an important role in multiple cancers, its role and regulatory mechanism in CRC is unclear. In this study, we aimed to investigate the role and clinical significance of Mex3a in CRC and to explore its underlying mechanism.
Methods
Western blotting and quantitative real-time polymerase chain reaction (qRT-PCR) were performed to detect the expression levels of genes. 5-Ethynyl-2'-deoxyuridine (EDU) and transwell assays were utilized to examine CRC cell proliferation and metastatic ability. The R software was used to do hierarchical clustering analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Overexpression and rescue experiments which included U0126, a specific mitogen activated protein kinase kinase/extracellular regulated protein kinase (MEK/ERK) inhibitor, and PX-478, a hypoxia-inducible factor 1 subunit alpha (HIF-1α) inhibitor, were used to study the molecular mechanisms of Mex3a in CRC cells. Co-immunoprecipitation (Co-IP) assay was performed to detect the interaction between two proteins. Bioinformatics analysis including available public database and Starbase software (starbase.sysu.edu.cn) were used to evaluate the expression and prognostic significance of genes. TargetScan (www.targetscan.org) and the miRDB (mirdb.org) website were used to predict the combination site between microRNA and target mRNA. BALB/c nude mice were used to study the function of Mex3a and hsa-miR-6887-3p in vivo.
Results
Clinicopathological and immunohistochemical (IHC) studies of 101 CRC tissues and 79 normal tissues demonstrated that Mex3a was a significant prognostic factor for overall survival (OS) in CRC patients. Mex3a knockdown substantially inhibited the migration, invasion, and proliferation of CRC cells. Transcriptome analysis and mechanism verification showed that Mex3a regulated the RAP1 GTPase activating protein (RAP1GAP)/MEK/ERK/HIF-1α pathway. Furthermore, RAP1GAP was identified to interact with Mex3a in Co-IP experiments. Bioinformatics and dual-luciferase reporter experiments revealed that hsa-miR-6887-3p could bind to the 3'-untranslated regions (3'-UTR) of the Mex3a mRNA. hsa-miR-6887-3p downregulated Mex3a expression and inhibited the tumorigenesis of CRC both in vitro and in vivo.
Conclusions
Our study demonstrated that the hsa-miR-6887-3p/Mex3a/RAP1GAP signaling axis was a key regulator of CRC and Mex3a has the potential to be a new diagnostic marker and treatment target for CRC.
Abbreviations
-
- CRC
-
- colorectal cancer
-
- Mex3a
-
- Mex-3 RNA binding family member A
-
- miRNA
-
- MicroRNA
-
- RAP1GAP
-
- RAP1 GTPase activating protein
-
- IHC
-
- immunohistochemistry
-
- TCGA
-
- The Cancer Genome Atlas
-
- GEO
-
- gene expression omnibus
-
- OS
-
- overall survival
-
- RFS
-
- recurrence-free survival
-
- RBP
-
- RNA binding protein
-
- UCSC
-
- University of California Santa Cruz
-
- SEM
-
- standard error of the mean
-
- ANOVA
-
- analysis of variance
-
- EDU
-
- 5-Ethynyl-2'-deoxyuridine
-
- UTR
-
- untranslated regions
-
- PMSF
-
- phenylmethanesulfonyl fluoride
-
- ECM
-
- endothelial cell medium
-
- DMEM
-
- dulbecco's modified eagle medium
-
- RPMI
-
- roswell park memorial institute
-
- FBS
-
- fetal bovine serum
-
- CCK8
-
- cell counting kit 8
-
- SDS
-
- sodium dodecyl sulfate
-
- GAPDH
-
- glyceraldehyde-3-phosphate dehydrogenase
-
- MOI
-
- multiplicity of infection
-
- 5-FU
-
- 5-fluorouracil
-
- IgG
-
- immunoglobulin G
-
- MEK
-
- mitogen activated protein kinase kinase
-
- p-MEK
-
- phosphor-mitogen activated protein kinase kinase
-
- ERK
-
- extracellular regulated protein kinase
-
- p-ERK
-
- phosphor-extracellular regulated protein kinase
-
- KEGG
-
- Kyoto Encyclopedia of Genes and Genomes
-
- MAPK
-
- mitogen-activated protein kinase
-
- HIF-1α
-
- hypoxia-inducible factor 1 subunit alpha
-
- Co-IP
-
- Co-immunoprecipitation
-
- KH
-
- K homology
-
- HUVEC
-
- human umbilical vein endothelial cell
-
- IRS
-
- immunoreactive score
-
- EDU
-
- 5-Ethynyl-2'-deoxyuridine
-
- BCA
-
- bicinchoninic acid
-
- HRP
-
- horseradish peroxidase
-
- qRT-PCR
-
- Quantitative real-time polymerase chain reaction
-
- AJCC
-
- American Joint Committee on Cancer
1 BACKGROUND
CRC is ranked third in incidence and second for mortality among malignant tumors globally [1]. To date, the most effective way to treat CRC is radical surgery combined with chemotherapy [2]. Although chemotherapy drugs have significantly improved the outcome of CRC patients in late stages, only 15%-25% of the patients are responsive to the treatments. For patients who can undergo surgery, up to 20% develop locoregional or distant recurrences within 5 years after surgery [3]. Therefore, discovery of new oncogenes is of great clinical significance to better decipher CRC pathophysiology and provide new therapeutics.
Mex3a was originally found in C. elegans as a translation-regulating protein that helps to maintain the totipotency of the reproductive line [4]. In humans, the Mex3 family has four homologous proteins Mex3a, b, c, and d [4]. Previous studies have recognized the critical oncogenic role of Mex3a in lung [5], gastric [6], breast [7], liver cancers [8], bladder urothelial carcinoma [9], Wilm's tumor [10], and glioblastoma multiforme [11]. Mex3a has also been reported to be involved in CRC and related with “stemness”, which means that it has unlimited proliferative potential [12]. However, its underlying mechanisms and regulation in CRC remained poorly understood.
MicroRNAs (miRNA) are short-chain non-coding RNAs consisting of 19-23 nucleotides. miRNAs inhibit target gene expression by sequence-specific interaction with the 3'- UTR, resulting in mRNA degradation and translation inhibition [13]. miRNAs play important roles in the processes of cancer development, such as differentiation, invasion, proliferation, and apoptosis [14-16]. In CRC, an increasing number of miRNAs have been implicated in tumorigenesis and were proposed as markers of prognosis or cancer therapy [17]. For instance, miR-214 was shown to inhibit CRC growth and liver metastasis by regulating fibroblast growth factor receptor 1 (FGFR1)[18]. Therefore, we hypothesized that a certain miRNA could be critical for the regulation of Mex3a in CRC.
Herein, in this study, we aimed to investigate the expression and effect of Mex3a in CRC, and further explored the underlying upstream and downstream mechanism to offer novel insight into CRC pathogenesis.
2 MATERIALS AND METHODS
2.1 Cell lines and cell culture
Human CRC cell lines SW620, HCT116, HT-29, SW480, Caco-2, normal colon cell line CCD841, HUVECs, and HEK293a were purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, P.R. China). HCT116 and HEK293a cells were cultured in regular roswell park memorial institute (RPMI) 1640 medium (#CF0001, SparkJade, Shandong, P.R. China). HUVECs were cultured in ECM (#1001, ScienCell, New York, USA) medium. Other cell lines were cultured in complete dulbecco's modified eagle medium (DMEM) (Gibco, Grand Island, NY, USA). All cell cultures were supplemented with a mixture of 10% fetal bovine serum (FBS) (#04-001-1A, Biological Industries, Hubei, China) and 1% penicillin/streptomycin (#BL505A, Biosharp, Shanghai, China). All cells were incubated at 37°C in a 5% CO2 atmosphere.
2.2 CRC patient samples and immunohistochemistry
Human CRC microarray (training cohort) for Mex3a staining were purchased from Shanghai Outdo Biotech, China. A validation cohort for Mex3a staining consisted of CRC and para-tumor tissues that were surgically resected from CRC patients at Qilu Hospital (Jinan, Shandong, China) between September 2017 and October 2019. Human CRC for RAP1GAP staining were also surgically resected from Qilu Hospital between September 2017 and October 2019. All diagnoses were confirmed by pathology. All experiments were approved and supervised by the Medical Ethics Committee of Qilu Hospital of Shandong University. TNM stages were classified according to the eighth American Joint Committee on Cancer (AJCC) edition. Immunohistochemistry was performed according to standard protocols using Mex3a and RAP1GAP antibodies from Abcam (Cambridge, UK). The expression levels of Mex3a and RAP1GAP were scored using the IHC scoring method [19] by a pathologist (KH) and an investigator (HXL). For Table 1–3, we divided patients into high expression or low expression groups according to IHC score.
| Factor | Total (cases [%]) | Mex3a expression (cases) | ||
|---|---|---|---|---|
| Low | High | |||
| Total | 101 | 53 | 48 | P valuea |
| Age | 0.134 | |||
| <65 years | 50 (49.5) | 30 | 20 | |
| ≥65 years | 51 (50.5) | 23 | 28 | |
| Gender | 0.392 | |||
| Male | 46 (45.5) | 22 | 24 | |
| Female | 55 (54.5) | 31 | 24 | |
| pT status | 0.025 | |||
| T1 | 1 (0.9) | 0 | 1 | |
| T2 | 10 (9.9) | 7 | 3 | |
| T3 | 61 (67.3) | 37 | 24 | |
| T4 | 29 (28.7) | 9 | 20 | |
| pN status | 0.919 | |||
| N0 | 61 (60.4) | 32 | 29 | |
| N1 | 30 (29.7) | 17 | 13 | |
| N2 | 10 (9.9) | 4 | 6 | |
| pM status | 1 | |||
| M0 | 100 (99.0) | 53 | 47 | |
| M1 | 1 (1.0) | 0 | 1 | |
| pTNM stageb | 0.325 | |||
| I | 6 (5.9) | 5 | 1 | |
| II | 55 (54.5) | 28 | 27 | |
| III | 39 (38.6) | 20 | 19 | |
| IV | 1 (1.0) | 0 | 1 | |
| Histopathological grade | 0.035 | |||
| Well | 23 (22.8) | 15 | 8 | |
| Moderate | 67 (66.3) | 36 | 31 | |
| Poor | 11 (10.9) | 2 | 9 | |
| Event | 0.012 | |||
| Alive | 49 (48.5) | 32 | 17 | |
| Dead | 52 (51.5) | 21 | 31 | |
- Abbreviations: CRC: colorectal cancer; Mex3a: Mex-3 RNA binding family member A.
- a The Pearson chi-squared test or Fisher's exact test was used for statistical analysis
- b TNM stages were classified according to the eighth edition of the American Joint Committee on Cancer (AJCC) staging system
2.3 EDU assay
All transfected with siRNA, plasmid, lentivirus, miR-6887-3p mimics or miR-6887-3p inhibitor cells were seeded into 24-well plates at 1 × 105 cells/well and incubated for 24 h. The proliferation ability was evaluated with an EDU assay kit (C10310-1, RIBO Biotechnology, Guangzhou, Guangdong, China) according to the manufacturer's instructions. Images were taken under a microscope (Olympus, Shanghai, China), and the percentage of EDU-positive cells was calculated using the formula EDU-positive cell count/total cell count × 100%.
2.4 Migration and Matrigel invasion assays
For migration and Matrigel invasion assays, 5000 cells were seeded in the upper chamber with 300 µL serum-free medium, and 600 µL medium containing 10% FBS were added to the lower chamber, which acted as a chemoattractant. After incubation for 48 h, the cells were wiped off the upper chamber with a cotton swab, and the invaded cells to the underside of the membrane were fixed and stained with 0.1% crystal violet (#C0121; Beyotime, Shanghai, China), and counted in five randomly selected microscopic fields (200×) per filter.
2.5 Cell viability assays
For cell counting kit 8 (CCK8) assay (#BB-4221; BestBio, Nanjing, Jiangsu, China), SW620 cells transfected with Mex3a siRNA or HCT116 cells transfected with Mex3a overexpression plasmid were resuspended and counted, then cells were plated into 96-well plates at a density of 1 × 104 cells per well and were treated with 5-fluorouracil (5-FU) (50, 100, 250, 500, 1000, 2000, 3000 µmol/L) (#F6627, Sigma, Shanghai, China). After 24 h of 5-FU treatment, the cell viability was evaluated by assessing the optical density (OD) at 450 nm following the manufacturer's instructions.
2.6 Co-culture angiogenesis assay
First, precooled u-Dish 35mm, high Grid-500 ibiTreat were coated with 200 µL of Matrigel (#354248, Corning Technology, Shanghai, China) at 37°C for 30 min. HUVECs were seeded at 2.5 × 104 cells/well, SW620 and SW480 cells were transfected with Mex3a siRNA or HCT116 cells were transfected with Mex3a overexpression plasmid, after 24 h of transfection, CRC cell supernatants were added to the HUVECs dish at a volume of CRC cell supernatants: ECM equal to 1:1. Tube formation was captured under a microscope (Olympus), and the number of branches and tube lengths were calculated using ImageJ software (National Institutes of Health).
2.7 Western blotting analysis
Cells transfected with siRNA, plasmid, lentivirus, miR-6887-3p mimics or miR-6887-3p inhibitor were harvested in sample buffer supplemented with Protease/Phosphatase Inhibitor Cocktail (#5872, Cell Signaling Technology, Shanghai, P.R. China). Protein levels were detected and calculated using the bicinchoninic acid (BCA) assay (#AR0197A, BOSTER, Shanghai, P.R. China) according to the manufacturer's instructions. Equal amounts (20 µg) of protein were added into sodium dodecyl sulfate (SDS) polyacrylamide gel, and the gel was transferred to polyvinylidene difluoride membranes (#10600023, GE, Shanghai, P.R. China). The membrane was incubated with primary antibodies overnight after blocking by milk. Then, the membrane was incubated with second antibody to be photographed. All used antibodies were Mex3a (1:1000 dilution; #ab79046, Abcam), RAP1GAP (1:5000 dilution; #ab32373, Abcam), MEK (1:500 dilution; #8727, Cell Signaling Technology, Shanghai, P.R. China), p-MEK (1:500 dilution #9154, Cell Signaling Technology, Shanghai, P.R. China), ERK (1:1000 dilution; #4695, Cell Signaling Technology, Shanghai, P.R. China), p-ERK (1:1000 dilution; #4370, Cell Signaling Technology, Shanghai, P.R. China), HIF-1α (1:1000 dilution; #ab51608, Abcam, Cambridge, UK) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:5000 dilution; #2118, Cell Signaling Technology, Shanghai, P.R. China).
2.8 Co-immunoprecipitation (Co-IP) assays
Co-IP assays were performed according to standard protocols. When SW620 cells' fullness reached more than 90%, the cells were scraped off directly with a cell scraper using immunoprecipitation lysis buffer NP-40 (#P0013F, Beyotime Institute of Biotechnology, Shanghai, P.R. China) with phenylmethanesulfonyl fluoride (PMSF)and protease inhibitor. Then, the cells were centrifuged at 12,000 rpm for 15 min, and the supernatant was collected, followed by incubation with primary antibodies (RAP1GAP) or isotype immunoglobulin G (IgG) (#3900, Cell Signaling Technology, Shanghai, P.R. China), gentle rocking 2 h at 4°C. Then, 35 µL of protein A/G beads (#sc-2003, Santa cruz, Shanghai, China) was added to each immunoprecipitation mixture with gentle rocking overnight at 4°C. The next day, the mixtures were washed five times with cold 1×Co-IP buffer, and bound protein was denatured with 2× SDS sample buffer. The supernatants were collected and proceeded to SDS-PAGE Western blot analysis.
2.9 Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from CRC cell lines using TRIzol (#15596-026, Invitrogen, CA, USA) following the manufacturer's instructions. cDNA was synthesized using a reverse transcription kit (#QP056, Genecopoeia, Guangzhou, Guangdong, P.R. China). GAPDH and U6 were used as internal parameters, and relative mRNA expression levels were calculated by comparing Ct method. The primers used were as follows: Mex3a-forward-5′-TGGAGAACTAGGATGTTTCGGG-3′; Mex3a-reverse-5′-GAGGCAGAGTTGATCGAGAGC-3′; hGAPDH-forward-5′-GGAAATCGTGCGTGACATTAA-3′; hGAPDH-reverse-5′-AGGAAGGAAGGCTGGAAGAG-3′; miR-6887-3p:5′-GAUCCUCCUUUCACCUCCCCU-3'; U6: HmiRQP9001; Genecopoeia, Guangzhou, China
2.10 Establishment of transient transfected cell lines
The cells were transiently transfected with siRNAs, plasmids, hsa-miR-6887-3p mimics and hsa-miR-6887-3p inhibitor using jetPRIME transfection reagent (#CPT114, Polyplus, New York, USA) according to the manufacturer's instructions. The cells were harvested to evaluate the transfection efficiency after 48 h. The sequence of Mex3a siRNA, RAP1GAP siRNA, hsa-miR-6887-3p inhibitor, hsa-miR-6887-3p mimics and control are shown in Table S1. Plasmid sequence of Mex3a and RAP1GAP are shown in Table S2.
2.11 Establishment of stable Mex3a-overexpressing or -knockdown cell lines
Mex3a overexpression and knockdown lentiviruses were purchased from the Genecopia Company. Stable Mex3a-overexpressing HCT116 cell lines were generated by being infected with lentivirus multiplicity of infection (MOI): 5 and selected with 0.3 mg/mL puromycin (#540411, Sigma, Shanghai, China) for about 1 week. Stable Mex3a-knockdown SW620 and SW480 cell lines were generated by being infected with lentivirus (MOI: 50 and 20, respectively) and selected with 1 mg/mL puromycin for about 1 week. Finally, the stable cell lines were verified using Western blot.
2.12 In vivo tumor xenograft assays
Six-week-old male BALB/c nude mice were purchased from Vital River Laboratory Animal Technology Co, Ltd (Beijing, P.R. China). Animal experimental procedures were approved by the Medical Ethics Committee of Shandong University (ECAESDUSM 2014056). The six-week-old male mice were randomized into different groups. SW480, HCT116, and SW620 cells (5 × 106 cells/mice) were implanted subcutaneously into the flank of nude mice. After tumor formation, tumor volume (mm3) was measured every other day and calculated by the formula (length × width × width)/2. When the tumors had reached a volume of approximately 600 mm3, the mice were euthanized, and the tumors were excised and embedded in paraffin for IHC analysis.
2.13 Transcriptome sequencing and bioinformatics analysis
Genome-wide transcriptional sequencing was performed by Baimaike (Beijing, China). Transcriptome sequencing (NEB, USA) was used to identify mRNA transcripts with differential expression between Mex3a-knockdown SW480 cells and control SW480 cells. The RNA preparation and library preparation for transcriptome sequencing were performed according to the manufacturer's instructions.
The gene expression profiles and clinical data of CRC patients from the The Cancer Genome Atlas (TCGA) were downloaded from the University of California Santa Cruz (UCSC) Xena Browser (https://xenabrowser.net/). The expression and OS analysis of miR-6887-3p was downloaded from the Starbase software (starbase.sysu.edu.cn). The gene expression and clinical data of CRC patients from the gene expression omnibus (GEO) datasets GSE39582 were obtained (https://www.ncbi.nlm.nih.gov/gds). R software was used to do hierarchical clustering analysis and KEGG pathway analysis.
2.14 Luciferase activity assay
The putative miRNA of Mex3a was predicted using TargetScan (www.targetscan.org) and miRDB (mirdb.org). For luciferase activity assay, the 3′ UTR region of the Mex3a gene was generated by standard polymerase chain reaction (PCR) techniques and inserted into the vector. A mutant type (MT) construct that disrupted the miR-6887-3p-binding sites of the Mex3a 3′ UTR segment was also amplified using a Quick change Site-Directed Mutagenesis Kit (#200518, Agilent, Shanghai, P.R. China). Wild type (WT) 3′ UTR or MT 3′ UTR plasmid of Mex3a was co-transfected with miR-6887-3p mimic into HEK293a cells using Lipofectamine 2000 (#11668-019, Invitrogen, Shanghai, P.R. China). Luciferase activity was detected by a Luciferase Reporter Gene Assay Kit (#GM-040501; Genomeditech, Shanghai, P.R. China) after transfected for 48 h.
2.15 Statistical analyses
All statistical analyses were performed using GraphPad Prism Software (version 6.0) or R software (version 3.6.1). Each assay was performed at least three independent replicates, and all data are presented as mean ± standard error of the mean (SEM). For comparisons, Student's t-test (two-sided), One-Way analysis of variance (ANOVA), Pearson's chi-square test, log-rank test, Kaplan–Meier survival analysis, Fisher's exact test, and Pearson's correlation analysis were performed as indicated. A P value < 0.05 was considered statistically significant.
3 RESULTS
3.1 Mex3a is highly expressed in CRC and correlates with poor prognosis
To clarify the role of Mex3a in CRC, we first analyzed Mex3a protein levels in a training cohort containing 101 CRC tissues and 79 normal tissues by IHC staining. Mex3a expression levels were found higher in tumor tissues than in adjacent normal tissues (Fig. 1A). IHC score indicated that Mex3a expression was significantly increased in CRC tissues compared to adjacent normal tissues (P < 0.001) (Fig. 1B). Kaplan-Meier analysis also revealed that patients with higher expression of Mex3a was associated with poorer OS (P = 0.003 (Fig. 1C). To confirm this finding, we detected Mex3a protein levels using Western blotting in eight randomly selected and paired CRC specimens. Six out of the eight specimens showed higher levels of Mex3a protein than the adjacent normal tissues (P = 0.049) (Fig. 1D).
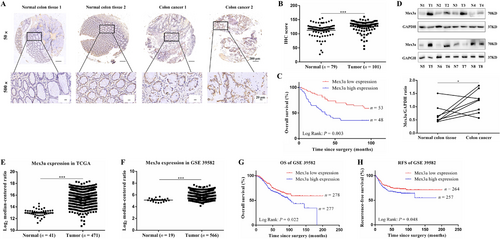
Mex3a is highly expressed in CRC and correlates with poor prognosis. A. IHC staining of Mex3a in the training cohort. Representative images of different Mex3a expression levels are shown, 50× scale bar 200 µm, 500× scale bar 20 µm. B. IHC scores of Mex3a expression in 79 normal tissues and 101 CRC tissues. C. Kaplan–Meier representation of the survival probability of the two groups of patients with high (n = 48) or low (n = 53) Mex3a expression in CRC tissues. Statistical analyses were performed with the log-rank test. D. Mex3a protein level is higher in tumor than in peritumoral tissues, as determined by Western blot. E. the mRNA expression levels of Mex3a in TCGA is obtained from Oncomine. F. the mRNA expression levels of Mex3a are obtained from the GEO dataset (GSE 39582). G. Kaplan–Meier representation of the OS of the two groups of patients with high (n = 277) or low (n = 278) Mex3a expression in CRC tissues. Statistical analyses were performed with the log-rank test. H. Kaplan–Meier representation of the RFS of the two groups of patients with high (n = 257) or low (n = 264) Mex3a expression in CRC tissues. Statistical analyses were performed with the log-rank test.
Data are shown as mean ± SEM; * P < 0.05; *** P < 0.001. Abbreviations: CRC: colorectal cancer; IHC: immunochemistry; TCGA: The Cancer Genome Atlas; GEO: gene expression omnibus; OS: overall survival; RFS: recurrence-free survival.
Next, we analyzed the mRNA level of Mex3a in human CRC samples from the TCGA and GEO dataset (GSE39582). Our results showed that the Mex3a expression level was significantly higher in CRC tissues than in normal tissues (P < 0.001) (Fig. 1E, F). We then investigated the relationship between Mex3a expression and survival characteristics in 566 CRC patients from the GSE39582 dataset. The results revealed that patients with higher Mex3a expression levels had poorer OS and recurrence-free survival (RFS) (Fig. 1G, 1H). These results suggest that Mex3a was upregulated in CRC and was closely related to its poor prognosis.
We further investigated the relationship between Mex3a and the clinicopathological features of CRC. We divided 101 CRC patients into Mex3a low-expression (52.5%, 53/101) and high-expression (47.5%, 48/101) groups according to their IHC score. We found that high expression of Mex3a was positively correlated with histopathological grade and tumor pathological T stage (pT stage) (P = 0.025, Table 1), while there was no correlation between Mex3a expression and sex, age, pN stage, pM stage, and pTNM stage. These results further confirmed that Mex3a high expression in CRC was correlated with poor prognosis.
3.2 High expression of Mex3a promotes tumorigenic properties of CRC cells
To investigate the potential roles of Mex3a in the development of CRC, we first measured Mex3a expression levels in five CRC cell lines compared to one normal colon cell line (CCD841). Among the five CRC cell lines, SW620, SW480, and Caco-2 cells showed the highest Mex3a expression at mRNA and protein levels, while HCT116 and HT-29 cells showed similar Mex3a levels as normal control CCD841 cells (Fig. S1A, S1B). Therefore, SW620 and SW480 were chosen for Mex3a silencing, and HCT116 was chosen for Mex3a overexpression. The efficiencies of Mex3a silencing and Mex3a overexpression were validated by Western blotting (Fig. 2A and 2B).
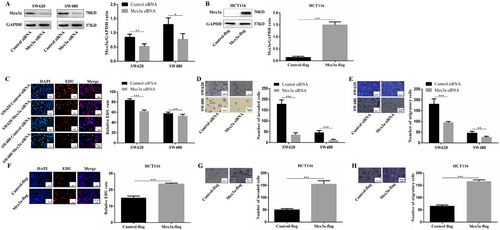
High expression of Mex3a promotes tumorigenic properties of CRC cells. A. Whole-cell lysates from SW620 and SW480 CRC cells instantaneously transduced with the siRNA (Control siRNA and Mex3a siRNA) are subjected to Western blotting analysis of Mex3a and GAPDH (as a loading control). Data are representative of four individual experiments. B. Whole-cell lysates from HCT116 CRC cells instantaneously transduced with the plasmid (Control-flag and Mex3a-flag) are subjected to Western blotting analysis of Mex3a and GAPDH (as a loading control). Data are representative of four individual experiments. C. EDU assays are performed in SW620 and SW480 cells with different levels of Mex3a expression. Scale bar 20 µm. D. Matrigel invasion assays shows the effect of Mex3a expression levels on invasion abilities of SW620 and SW480 cells. Scale bar 50 µm. E. Migration assays shows the effect of Mex3a expression levels on migratory abilities of SW620 and SW480 cells. Scale bar 50 µm. F. Representative EDU assay shows that high level of Mex3a induced proliferation of HCT116 CRC lines. scale bar 20 µm. G. Representative matrigel invasion assay shows that high level of Mex3a induced invasion of HCT116 CRC lines. Scale bar 50 µm. H. Migration assays shows that a high level of Mex3a induced migratory of HCT116 cells. Scale bar 50 µm.
Data are shown as mean ± SEM; * P < 0.05; ** P < 0.01; *** P < 0.001. Abbreviations: CRC: colorectal cancer; EDU: 5-Ethynyl-2'-deoxyuridine.
Next, SW640 and SW480 were used as the cell models for RNA knockdown. EDU assay revealed that Mex3a knockdown significantly impaired CRC cell proliferation (Fig. 2C). Transwell assays were performed to detect the effects of Mex3a on cell invasion and migration. Our results revealed that Mex3a knockdown substantially attenuated the metastatic ability of SW620 and SW480 cells (Fig. 2D, E).
Angiogenesis experiments were also performed to detect the effect of Mex3a on angiogenesis regulation. SW620 and SW480 cells were transfected with Mex3a siRNA or Control siRNA. Their supernatants were then collected and used to treat HUVECs. Mex3a silencing was found to reduce the number of branches and tubule length of HUVECs compared to those of the control groups (Fig. S1C). Finally, over a range of drug concentrations, Mex3a knockdown significantly enhanced sensitivity to 5-FU in SW620 cells (Fig. S1D). As expected, overexpression of Mex3a in HCT116 cells resulted in the opposite results (Fig. 2F-H and Fig. S1E, S1F). These results indicate that Mex3a could promote proliferation, invasion, and angiogenesis in CRC cells in vitro.
3.3 Mex3a enhances oncogenesis of CRC cells in vivo
To investigate the functional role of Mex3a in vivo, we constructed lentivirus-mediated Mex3a knockdown SW480 cells and SW620 cells, and Mex3a overexpression HCT116 cells (Fig. 3A, B). Thereafter, SW480 and HCT116 cells with stable knockdown or overexpression of Mex3a were transplanted into nude mice. Knockdown of Mex3a in SW480 slowed the in vivo tumor growth (Fig. 3C-E). IHC staining showed that the staining intensity and expression levels of Ki-67 were both decreased in the Mex3a knockdown group compared to normal tissues (Fig. S1G). Moreover, xenotransplant experiments showed that enforced Mex3a expression markedly increased the tumorigenicity of HCT116 cells (Fig. 3F-H). IHC staining showed that the staining intensity and expression levels of Ki-67 were both increased in the Mex3a overexpression groups (Fig. S1H). Collectively, these results indicate that Mex3a could promote oncogenesis in nude mice.
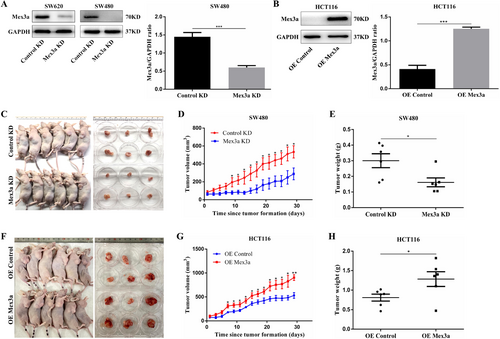
Mex3a enhances oncogenesis of CRC in vivo. A. whole-cell lysates from SW620 and SW480 CRC cells stably transduced were subjected to Western blotting analysis of Mex3a and GAPDH (as a loading control). Data are representative of three individual experiments. B. Whole-cell lysates from HCT116 cells stably transduced are subjected to Western blotting analysis of Mex3a and GAPDH (as a loading control). Data are representative of four individual experiments. C. Effect of Mex3a knockdown on CRC tumorigenesis in vivo. D. The volume of subcutaneous tumors is measured (n = 6). E. The weight of subcutaneous tumors is measured (n = 6). F. Effect of Mex3a overexpression on CRC tumorigenesis in vivo. G. Measurement of the volume of subcutaneous tumors of each group (n = 6). H. Measurement of the weight of subcutaneous tumors of each group (n = 6).
Data are shown as mean ± SEM; * P < 0.05; ** P < 0.01; *** P < 0.001. Abbreviations: CRC: colorectal cancer.
3.4 Mex3a acts as a co-activator of RAP1GAP/MEK/ERK/HIF-1α signaling in CRC cells
To explore the possible mechanisms accounting for the role of Mex3a in the development of CRC, we performed transcriptome sequencing with stably transduced cell lines (SW480 control-knockdown and SW480 Mex3a-knockdown) to identify the relevant signaling pathway. Hierarchical clustering analysis showed that Mex3a knockdown led to a clear and consistent difference in gene expression profile (Fig. 4A). We identified 752 upregulated genes and 352 downregulated genes (|fold change| > 1, data not shown). The KEGG pathway analysis showed that Mex3a knockdown led to the enrichment of the mitogen-activated protein kinase (MAPK) and Rap1 pathway (Fig. 4B). Considering the differentially expressed genes involved in this pathway, we speculated that Mex3a could affect colon cancer phenotype by regulating the RAP1GAP/MEK/ERK/HIF-1α pathway.
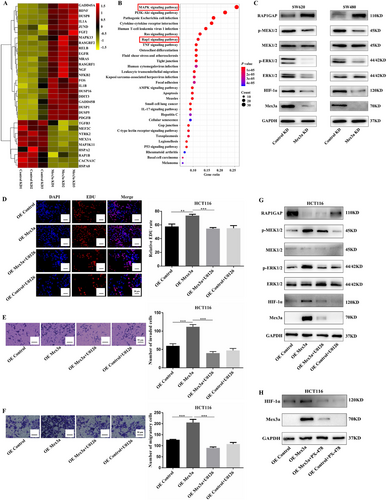
Mex3a acts as a co-activator of RAP1GAP/MEK/ERK/HIF-1α signaling in CRC cells. A. Heatmap of distinctly dysregulated mRNAs in stable SW480 cell lines (Control-knockdown vs Mex3a- knockdown) are identified from transcriptome sequencing by hierarchical clustering. High and low expression levels are indicated in red and yellow, respectively. B. Pathway analysis of distinctly dysregulated pathways in stable SW480 cell lines identified from exon and transcriptome sequencing. Statistical significance is indicated by different colors. The size of the circle represents the number of genes in each pathway. C. RAP1GAP/MEK/ERK/HIF-1α expression in SW620 and SW480 cells is detected by Western blot after Mex3a knockdown. D, E, F. EDU, Matrigel invasion and migration assay of the rescue experiment using U0126 (20 µmol/L) in HCT116 cells. Scale bar 20 µm and 50 µm, respectively. G. RAP1GAP/MEK/ERK/HIF-1α expression in HCT116 OE Mex3a cells was detected by Western blot after 20 µmol/L U0126 incubated for 24 h. H. HIF-1α expression in HCT116 OE Mex3a cells is detected by Western blot after 25 µmol/L PX-478 incubated for 24 h.
Data are shown as mean ± SEM; ** P < 0.01; *** P < 0.001. Abbreviations: CRC: colorectal cancer; EDU: 5-Ethynyl-2'-deoxyuridine; RAP1GAP: RAP1 GTPase activating protein; KEGG: Kyoto Encyclopedia of Genes and Genomes; MAPK: mitogen-activated protein kinase; HIF-1α: hypoxia-inducible factor 1 subunit alpha; MEK: mitogen activated protein kinase kinase; ERK: extracellular regulated protein kinase.
Rap1 is a small GTP-binding protein that cycles between the GTP-bound and GDP-bound states [20, 21]. RAP1GAP is lost in several tumor types, and thus, it may function as a tumor suppressor [22]. Overexpression of RAP1GAP has been reported to induce apoptosis [23], suppress cell proliferation [24, 25], inhibit cell cycle progression [26], and reduce tumor progression [27, 28]. Thus, RAP1GAP possesses most of the well-studied characteristics found in tumor-inhibiting factors, suggesting that RAP1GAP may play a role in the progression of CRC.
We, therefore, verified the expression of the proteins involved in the RAP1GAP signaling pathway in lentivirus-mediated Mex3a knockdown SW620 and SW480 cells by Western blotting. We found that in both cell lines, knockdown of Mex3a induced the upregulation of RAP1GAP and downregulation of phosphor-mitogen activated protein kinase kinase 1/2 (p-MEK1/2), phosphor-extracellular regulated protein kinase 1/2 (p-ERK1/2), and HIF-1α (Fig. 4C). We further performed a rescue experiment using U0126, a specific MEK/ERK inhibitor, and PX-478, an HIF-1α inhibitor. We discovered that using U0126 (20 µmol/L) partially attenuated the proliferation, invasion, and migration of HCT116 cells caused by Mex3a overexpression (Fig. 4D-F). In addition, U0126 reduced the expression of p-MEK1/2 and p-ERK1/2 and impaired the expression of HIF-1α induced by Mex3a overexpression (Fig. 4G). PX-478 (25 µmol/L) also inhibited HIF-1α expression induced by Mex3a overexpression (Fig. 4H). Interestingly, Mex3a expression was also reduced following U0126 and PX-478 treatment. Possibly, this is because the inhibition of these pathways altered the stability of the Mex3a protein through a feedback loop. These results indicated that Mex3a regulated tumor growth through the RAP1GAP/MEK/ERK/HIF-1α signaling in CRC cells.
3.5 Mex3a physically interacted with RAP1GAP in CRC cells
To elucidate the molecular mechanism by which Mex3a promotes tumorigenic properties in CRC cells, we next identified proteins that interact with Mex3a in CRC cells. Several lines of evidence have revealed that Mex3b could bind to RAP1GAP [29, 30]. Mex3a and Mex3b are Mex3 family homologous proteins that have the same functional domain. Therefore, we determined whether Mex3a interacts with RAP1GAP. Co-IP was used to confirm the protein association between Mex3a and RAP1GAP in SW620 and SW480 cells (Fig. 5A). Furthermore, we analyzed Mex3a protein levels in validation cohorts containing 100 cases of CRC and 6 normal tissues by IHC staining. The results showed that high expression of Mex3a was accompanied by low expression of RAP1GAP in tumor tissues (Fig. 5B). Similar to the training cohort, the IHC score indicated that Mex3a expression was significantly increased in CRC tissues compared with normal tissues (P < 0.001) (Fig. 5C). RAP1GAP expression was also significantly decreased in CRC tissues compared to normal tissues (P = 0.026) (Fig. 5D). The available data suggested that the protein level of Mex3a was negatively correlated with RAP1GAP (r = -0.235; P = 0.020) (Fig. 5E).
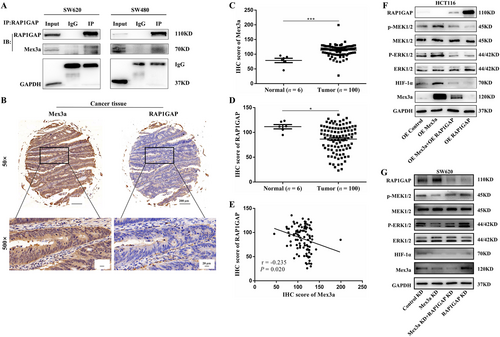
Mex3a physically interacted with RAP1GAP in CRC cells. A. The interaction between endogenous RAP1GAP and Mex3a was detected by immunoprecipitation in SW620 and SW480 cells. B. IHC staining of Mex3a and RAP1GAP expression in the validation cohort (6 cases of normal tissues and 100 cases of CRC). Representative images of different Mex3a and RAP1GAP expression levels are shown, 50×scale bar 200 µm, 500×scale bar 20 µm. C. IHC scores of Mex3a expression in the validation cohort. D. IHC scores of RAP1GAP expression. E. RAP1GAP are inversely correlated with Mex3a in CRC patients (r = -0.235; P = 0.020). F. RAP1GAP/MEK/ERK/HIF-1α expression in HCT116 cells was detected by Western blot after RAP1GAP overexpression. G. RAP1GAP/MEK/ERK/HIF-1α expression in SW620 cells is detected by Western blot after knockdown of RAP1GAP.
Data are shown as mean ± SEM; * P < 0.05; *** P < 0.001. Abbreviations: CRC: colorectal cancer; IHC: immunochemistry; RAP1GAP: RAP1 GTPase activating protein; Co-IP: Co-immunoprecipitation; HIF-1α: hypoxia-inducible factor 1 subunit alpha; MEK: mitogen activated protein kinase kinase; ERK: extracellular regulated protein kinase; IgG: immunoglobulin G.
Growing evidence suggests that the overexpression of RAP1GAP in human colon cancer cells impairs cell migration, invasion, and cell-matrix adhesion in vitro[31, 32]. We then performed a rescue experiment to explore whether Mex3a promoted the expression of MEK/ERK/HIF-1α through RAP1GAP. We found that RAP1GAP partially reduced the expression of p-MEK, p-ERK, and HIF-1α induced by Mex3a overexpression and that RAP1GAP knockdown reversed this decrease caused by Mex3a knockdown (Fig. 5F, G). Interestingly, Western blotting and qPCR showed that the RAP1GAP protein level was decreased but mRNA did not change after overexpression of Mex3a (Fig. 4G and Fig. S2A). These results indicated that RAP1GAP was lowly expressed in CRC and physically interacted with Mex3a in CRC cells.
We further investigated the role of RAP1GAP in CRC. First, we analyzed the mRNA level of RAP1GAP in human CRC samples from TCGA. The results showed that the RAP1GAP expression level was significantly lower in CRC tissues than in normal tissues (P < 0.001) (Fig. S2B). We then investigated the relationship between RAP1GAP expression and survival characteristics in 299 CRC patients from the TCGA. Our results revealed that lower RAP1GAP expression levels were associated with poorer OS (Fig. S2C). Moreover, we explored the relationship between the protein level of RAP1GAP and the clinicopathological features of CRC through tissue microarray. We divided 100 CRC patients into RAP1GAP low-expression (50%, 50/100) and high-expression (50%, 50/100) groups according to IHC score. We found that there was no correlation between high RAP1GAP expression and age, pT, pN, pM, pTNM stage, and histopathological grade except for gender (Table 2). In addition, the validation cohort confirmed that high Mex3a expression was positively correlated with pT stage (P = 0.043) and pN stage (P = 0.033) (Table 3). These results provided an insight that RAP1GAP may play an inhibitory role in CRC.
| RAP1GAP expression (cases) | ||||
|---|---|---|---|---|
| Factors | Total (cases [%]) | Low | High | |
| Total | 100 | 50 | 50 | P valuea |
| Age | 0.317 | |||
| <55 | 50 (50.0) | 28 | 22 | |
| ≥55 | 50 (50.0) | 22 | 28 | |
| Gender | 0.03 | |||
| Male | 69 (69.0) | 29 | 40 | |
| Female | 31 (31.0) | 21 | 10 | |
| pT status | 0.560 | |||
| T1 | 2 (2.0) | 2 | 0 | |
| T2 | 24 (24.0) | 12 | 12 | |
| T3 | 49 (49.0) | 24 | 25 | |
| T4 | 25 (25.0) | 12 | 13 | |
| pN status | 0.685 | |||
| N0 | 46 (46.0) | 24 | 22 | |
| N1 | 34 (34.0) | 15 | 19 | |
| N2 | 20 (20.0) | 11 | 9 | |
| pM status | 0.617 | |||
| M0 | 96 (96.0) | 49 | 47 | |
| M1 | 4 (4.0) | 1 | 3 | |
| pTNM stageb | 0.069 | |||
| I | 15 (15.0) | 3 | 12 | |
| II | 32 (32.0) | 18 | 14 | |
| III | 49 (49.0) | 26 | 23 | |
| IV | 4 (4.0) | 3 | 1 | |
| Histopathological grade | 0.623 | |||
| Well | 19 (19.0) | 8 | 11 | |
| Moderately | 62 (62.0) | 31 | 31 | |
| Poorly | 19 (19.0) | 11 | 8 | |
- Abbreviations: CRC: colorectal cancer; RAP1GAP: RAP1 GTPase activating protein
- a The Pearson chi-squared test or Fisher's exact test was used for statistical analysis
- b TNM stages were established according to the AJCC (American Joint Committee on Cancer) eighth edition.
| Mex3a expression (cases) | ||||
|---|---|---|---|---|
| Factors | Total (cases [%]) | Low | High | P valuea |
| Age | 0.682 | |||
| <55 | 50 (50.0) | 32 | 18 | |
| ≥55 | 50 (50.0) | 18 | 32 | |
| Gender | 0.194 | |||
| Male | 69 (69.0) | 31 | 38 | |
| Female | 31 (31.0) | 19 | 12 | |
| pT status | 0.043 | |||
| T1 | 2 (2.0) | 2 | 0 | |
| T2 | 24 (24.0) | 17 | 7 | |
| T3 | 49 (49.0) | 21 | 28 | |
| T4 | 25 (25.0) | 10 | 15 | |
| pN status | 0.033 | |||
| N0 | 46 (46.0) | 29 | 17 | |
| N1 | 34 (34.0) | 15 | 19 | |
| N2 | 20 (20.0) | 6 | 14 | |
| pM status | 0.617 | |||
| M0 | 96 (96.0) | 49 | 47 | |
| M1 | 4 (4.0) | 1 | 3 | |
| pTNM stageb | 0.655 | |||
| I | 15 (15.0) | 9 | 6 | |
| II | 32 (32.0) | 16 | 16 | |
| III | 49 (49.0) | 24 | 25 | |
| IV | 4 (4.0) | 1 | 3 | |
| Histopathological grade | 0.12 | |||
| Well | 19 (19.0) | 12 | 7 | |
| Moderately | 62 (62.0) | 26 | 36 | |
| Poorly | 19 (19.0) | 12 | 7 | |
- Abbreviations: CRC: colorectal cancer; Mex3a: Mex-3 RNA binding family member A.
- a The Pearson chi-squared test or Fisher's exact test was used for statistical analysis
- b TNM stages were established according to the AJCC (American Joint Committee on Cancer) eighth edition.
3.6 Mex3a is a target of hsa-miR-6887-3p in CRC cells
Now that we identified the oncogenesis role of Mex3a, we decided to find out the upstream regulatory mechanism of Mex3a. miRNAs are known to function by regulating target genes. The putative miRNA of Mex3a was predicted using TargetScan (www.targetscan.org) and miRDB (mirdb.org). The results showed that the 3′ UTR region of Mex3a possessed potential complementary sites for the miR-6887-3p seed region (Fig. 6A). A luciferase reporter assay was performed to further investigate the direct binding and inhibitory effects of miR-6887-3p and Mex3a. The Mex3a 3′ UTR-WT and corresponding MT were amplified and cloned downstream of a luciferase reporter gene in the pGL3 basic vector. In HEK293a cells, co-transfection of pcDNAmiR-6887-3p and Mex3a-3′ UTR-WT reduced luciferase activity compared with the Mex3a-3′ UTR-WT and mimics NC groups. However, no difference in luciferase activity was observed in cells co-transfected with Mex3a-3′ UTR-MT and pcDNAmiR-6887-3p (Fig. 6B). These results indicated that Mex3a was a target of hsa-miR-6887-3p in CRC cells. Analysis using the Starbase software (starbase.sysu.edu.cn) revealed that miR-6887-3p was under-expressed in colon cancer compared to the normal group (Fig. S2D). The high expression of miR-6887-3p correlated with high OS rate (Fig. S2E).
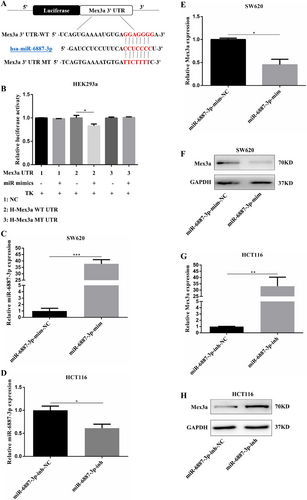
Mex3a is a target of hsa-miR-6887-3p in CRC Cells. A. The binding sites of miR-6887-3p in the Mex3a 3′ untranslated region. B.The pcDNAmiR-6887-3p expression plasmid or the negative control, a pGL3 luciferase vector containing the WT or MT of Mex3a 3′ UTR, were co-transfected into HEK293a cells, and the relative firefly luciferase activity is measured. The data are presented as the mean ± SEM of three independent experiments. C. Relative expression levels of miR-6887-3p in SW620 cells after transfection with miR-6887-3p mimics. D. Relative expression levels of miR-6887-3p in HCT116 cells after transfection with miR-6887-3p inhibitor. E, F. The Mex3a mRNA and protein levels in SW620 cells transfected with miR-6887-3p mimics or negative control (mim-NC) vector. G, H. The Mex3a mRNA and protein levels in HCT116 cells transfected with miR-6887-3p inhibitor or negative control (inh-NC) vector.
Data are shown as mean ± SEM; * P < 0.05; ** P < 0.01; *** P < 0.001. Abbreviations: CRC: colorectal cancer; UTR: untranslated regions; WT: wild type; MT: mutant type; qRT-PCR: quantitative real-time polymerase chain reaction.
To explore whether miR-6887-3p negatively regulated Mex3a expression in the CRC cellular environment, we first measured the expression level of miR-6887-3p in CRC cell lines and a normal colon cell line, CCD841. Among the three CRC cell lines, SW620 presented the lowest miR-6887-3p expression levels while HCT116 had relatively high miR-6887-3p expression levels (Fig. S2F). Thus, we used SW620 cells to overexpress miR-6887-3p by transfection with miR-6887-3p mimics. As HCT116 cells possessed relatively high miR-6887-3p levels, we knocked-down the endogenous miR-6887-3p by treating them with an miR-6887-3p inhibitor in HCT116 cells. Transfection efficiency was confirmed by qRT-PCR (Fig. 6C, D). mRNA and protein levels of Mex3a were measured. The results showed that ectopic expression of miR-6887-3p significantly decreased the Mex3a mRNA and protein levels in SW620 cells (Fig. 6E, F), whereas miR-6887-3p knockdown increased the Mex3a mRNA and protein expression in HCT116 cells (Fig. 6G, H). These results suggested that Mex3a could be a target of hsa-miR-6887-3p in CRC cells.
3.7 hsa-miR-6887-3p represses Mex3a to inhibit CRC growth
The above results prompted us to examine whether miR-6887-3p could inhibit CRC growth by inhibiting Mex3a. We first assessed whether miR-6887-3p could reverse the effect of Mex3a in CRC cells.
In SW620 cells transfected with miR-6887-3p mimics, Western blotting validated that overexpression of miR-6887-3p by transfection with miR-6887-3p mimics suppressed Mex3a expression (Fig. 7A). Compared with the control group, SW620 cells transfected with miR-6887-3p mimics displayed notably reduced cell proliferation and invasion (Fig. 7B, C), This was similar to the result of the low expression of Mex3a. HCT116 cells transfected with the miR-6887-3p inhibitor showed opposite results (Fig. 7D-F). To examine whether the function of miR-6887-3p depended on the expression of Mex3a in CRC cells, SW620 cells were transduced with miR-6887-3p mimics and Mex3a overexpression plasmid. Interestingly, ectopic Mex3a expression could at least partially rescue the proliferation and invasion ability inhibited by miR-6887-3p (Fig. 7C, D). HCT116 cells were transduced with miR-6887-3p inhibitor and Mex3a siRNA. Mex3a knockdown also partially rescued the proliferation and invasion ability promoted by inhibition of the expression of miR-6887-3p (Fig. 7E, F). Altogether, these results showed that hsa-miR-6887-3p could inhibit the proliferation and invasion of CRC cells by suppressing Mex3a.
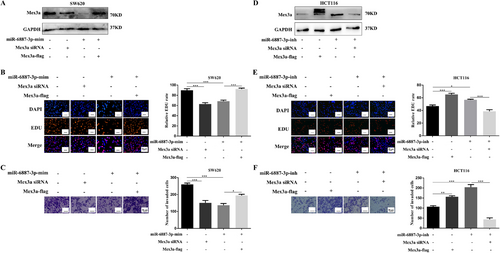
hsa-miR-6887-3p represses Mex3a to inhibit CRC growth. A. Western blot assays were performed to investigate the expression of Mex3a after miR-6887-3p overexpression, Mex3a knockdown, and Mex3a reintroduction in SW620 cells. B, C. Cell proliferation and invasion in SW620 cells after transfected with different plasmids. Scale bar 20 µm and 50 µm, respectively. D. Western blot assays are performed to investigate the expression of Mex3a after transfection with miR-6887-3p inhibitor, Mex3a overexpression, and Mex3a knockdown in HCT116 cells. E, F. Cell proliferation and invasion in HCT116 cells after transfection with different plasmids. Scale bar 20 µm and 50 µm, respectively.
Data are shown as mean ± SEM; * P < 0.05; ** P < 0.01; *** P < 0.001. Abbreviations: CRC: colorectal cancer; EDU: 5-Ethynyl-2'-deoxyuridine.
3.8 Ectopic expression of miR-6887-3p suppresses CRC cell growth in vivo
To investigate the role of miR-6887-3p in tumor growth in vivo, SW620 cells were stably transfected with LV-miR-6887-3p to overexpress miR-6887-3p. Transfection efficiency was confirmed by qRT-PCR (Fig. 8A). The transfected cells were then injected into the flanks of 6-week-old male nude mice. The tumor was measured every other day. The mice were euthanized, and the tumors were collected after 18 days. Our results showed that the tumor size of the SW620-Lv-miR-6887-3p group was significantly smaller than that of the SW620-Lv-miR-NC group (Fig. 8B-D). Moreover, in the SW620-Lv-miR-6887-3p group, the staining intensity and expression levels of Ki-67 were decreased (Fig. S2G). Taken together, these results indicate that miR-6887-3p could suppress tumorigenesis in nude mice.
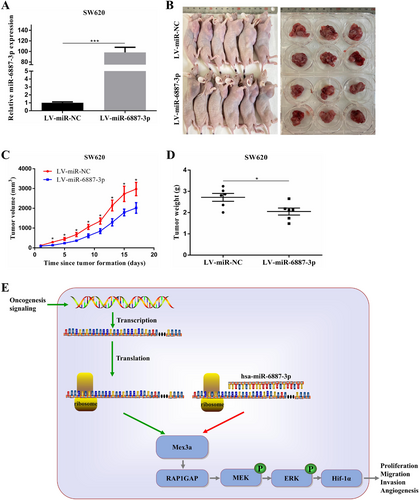
Ectopic expression of miR-6887-3p suppresses CRC cell growth in vivo. A. Relative expression levels of miR-6887-3p in SW620 cells transfected with LV-miR-6887-3p. B. Effect of miR-6887-3p overexpression in SW620 on CRC tumorigenesis in vivo. C. Measurement of the volume of subcutaneous tumors of each group (n = 6). D. Measurement of the weight of subcutaneous tumors of each group (n = 6). E. Diagram of hsa-miR-6887-3p-Mex3a axis on oncogenesis in CRC. After decreased expression of hsa-mir-6887-3p due to tumor-promoting signals, Mex3a expression is increased, leading to the downregulation of RAP1GAP to activate the MEK/ERK/ HIF-1α pathway. Ultimately this axis can promote CRC cell proliferation, invasion, migration, and angiogenesis as well as tumor formation.
Data are shown as mean ± SEM; * P < 0.05; *** P < 0.001. Abbreviations: CRC: colorectal cancer; RAP1GAP: RAP1 GTPase activating protein; qRT-PCR: quantitative real-time polymerase chain reaction.
4 DISCUSSION
The dysregulation of oncogenic genes is a key step in the development of cancers, including CRC. In order to develop innovative therapeutic strategies, new molecular participants responsible for CRC phenotypes must be identified and described. Here, we discovered that the RNA-binding ubiquitin ligase, Mex3a, was overexpressed in CRC tissue from public available datasets and the role of Mex3a in CRC was still unknown.
Our study highlights the function of Mex3a, one of the RNA binding proteins (RBPs), and its mechanisms in regulating CRC angiogenesis. Mex3a was identified as a novel presumptive oncogene in CRC. Previous studies have shown that Mex3a might be an oncogenic substance in gastric cancer [6], glioma [11], Wilms tumors [10], and bladder cancer [9]. Mex3a not only degrades its target mRNA as an RBP but also promotes ubiquitylation as a ubiquitin ligase [11, 12]. In this present study, we found that Mex3a was markedly upregulated in CRC cell lines and tissues, and overexpression of Mex3a promoted CRC cells proliferation, invasion and migration. In CRC tissues high Mex3a expression was also correlated with tumor size, pT stage, histological grade, and OS. Thus, Mex3a could be a strong predictor of CRC carcinogenesis and prognosis. Similarly, Yang et al. [8] reported that Mex3a was significantly associated with poor survival in liver cancer. Furthermore, we found that Mex3a inhibited the protein expression of RAP1GAP but had no effect on its mRNA level. Moreover, another study revealed that Mex3b and RAP1GAP physically interacted in an RNA-independent manner [29]. Human MEX3 proteins consist of two N-terminal K homology (KH) domains and a C-terminal RING finger module, which are responsible for the RNA-binding ability and ubiquitin E3-ligase activity, respectively[4]. Therefore, we hypothesized that the decreased protein level of RAP1GAP without mRNA change might be related to ubiquitination considering the role of Mex3a as a ubiquitin ligase. In the future, our laboratory will be devoted to the study whether Mex3a regulated the expression of RAP1GAP in a ubiquitin-dependent manner.
The regulation of signaling pathways in many tumors are complicated. RNA-seq revealed that the knockdown of Mex3a altered the Rap1 and MAPK signaling pathways. Previous studies have shown that RAP1GAP inhibits the activation of MEK/ERK signaling in many malignancies [33, 34]. Rap1/MEK/ERK signaling as a classical pathway promotes carcinogenesis [35, 36]. RAP1GAP, as a tumor suppressor, inhibits proliferation, migration [24, 25, 37], invasion [38, 39], and apoptosis [40] in many malignancies. In colon cancer, the depletion of RAP1GAP promoted migration and invasion [31]. Multiple studies have revealed that MEK/ERK facilitates the stabilization and transcriptional activity of HIF-1α [41, 42]. In our study, we detected Rap1 and MAPK pathway disturbance induced by Mex3a and found that Mex3a promoted proliferation and invasion of CRC through the RAP1GAP/MEK/ERK/HIF-1α signaling pathway. Moreover, inhibitors of MEK/ERK and HIF-1α reversed the oncogenic activity of Mex3a. Overexpression of RAP1GAP also reversed the oncogenic effect of Mex3a, that is to say, RAP1GAP physically interacts with Mex3a according to the results of Co-IP and thus inactivating the RAP1GAP/MEK/ERK/HIF-1α pathway.
Changes in miRNA expression levels have been reported in many tumors [43, 44]. They may act as either tumor suppressors or oncogenes in cancer. Expression profiles of miRNAs with biomarker potential can be used for the classification, diagnosis, targeted therapy, and prognosis of different cancer types. It is well known that miRNAs play a regulatory role in translation by targeting the 3′ UTR site of mRNA. Here, we identified Mex3a as a target of hsa-miR-6887-3p according to the results of luciferase reporter assays. Bioinformatic analyses confirmed that miR-6887-3p was downregulated in CRC tissues and low expression of miR-6887-3p was associated with poor OS. Gain-of-function or loss-of-function experiments suggested that miR-6887-3p inhibited the proliferation and invasion of CRC cells in vitro. Researchers have reported that miR-6887-3p inhibits proliferation and glycolytic metabolism in HESCs [45]. Considering that hsa-miR-6887-3p binds to the 3'-UTR of the Mex3a mRNA, the combination could inhibit the transcription of Mex3a. Thus, decreased hsa-miR-6887-3p expression can cause up-regulation of Mex3a. In the present study, we reported that miR-6887-3p might be a tumor-inhibiting factor that negatively regulates colon cancer by directly inhibiting the expression of Mex3a, thus inactivating the RAP1GAP/MEK/ERK/HIF-1α pathway.
In conclusion, the present study demonstrated that Mex3a and RAP1GAP played important roles in the development and progression of CRC. Meanwhile, Mex3a and RAP1GAP could interact with each other and activate the MEK/ERK/HIF-1α signaling pathway, which ultimately increased the invasion and proliferation of CRC cells. More importantly, our investigation revealed that high expression levels of Mex3a and low expression of RAP1GAP were significantly correlated with the progression of CRC and served as independent prognostic factors for the poor outcomes in CRC patients. Our results also suggested that miR-6887-3p markedly inhibited in vivo tumor formation, and inhibited the proliferation and invasion induced by Mex3a in CRC cells. The newly discovered hsa-miR-6887-3p/Mex3a and Mex3a/RAP1GAP/MEK/ERK/HIF-1α axis provide theoretical basis of pathogenesis of CRC and potential target therapy for the treatment of CRC.
DECLARATIONS
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHORSHIP
Conception and Design: CYL, HXL, KH.
Acquisition of Data: HXL, JHL, JW, SL.
Analysis and Interpretation of Data: HXL, JHL, JW, JYH.
Drafting the manuscript: HXL.
Revising the manuscript for intellectual content: HXL, JHL, KH, CYL.
Final approval of the completed manuscript: KH, CYL.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Informed consent was obtained from all patients. All experiments were approved and supervised by the Ethics Committee of Qilu Hospital of Shandong University and Ethics Committee of Shandong University.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
The study was supported by the National Scientific Foundation of China (NSFC32071127 and 31871160)
ACKNOWLEDGMENTS
We thank Dr. Wei Guo, Department of colorectal surgery, Qilu Hospital, Cheeloo College of Medicine, Shandong University, for providing the colorectal cancer tissues.
Open Research
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this published article.




