High-throughput sequencing detection and ensartinib treatment of lung cancer harboring NTRK1 fusion
Abbreviations
-
- NGS
-
- next-generation sequencing
-
- SNV
-
- single nucleotide variation
-
- indel
-
- insertion-or-deletion
-
- PANO-Seq
-
- parallel amplification and numerically optimized sequencing
-
- NTRK
-
- neurotrophic receptor tyrosine kinase
-
- LoD
-
- limit of detection
-
- MAF
-
- mutant allele frequency
-
- TKI
-
- tyrosine receptor kinase inhibitor
-
- EGFR
-
- epidermal growth factor receptor
-
- KRAS
-
- KRAS proto-oncogene
-
- PIK3CA
-
- phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
-
- BRAF
-
- B-Raf proto-oncogene, serine/threonine kinase
-
- MET
-
- MET proto-oncogene
-
- ALK
-
- anaplastic lymphoma kinase
-
- ROS1
-
- ROS proto-oncogene 1
-
- RET
-
- ret proto-oncogene
-
- NRG1
-
- neuregulin 1
-
- CHMP2A
-
- charged multivesicular body protein 2A
-
- HLA-DRB1
-
- human leukocyte antigen DRB1
-
- TPR
-
- translocated promoter region protein
-
- SQSTM1
-
- sequestosome 1
-
- RFWD2
-
- RING finger and WD repeat domain 2
-
- MSN
-
- moesin
-
- PR
-
- partial response
-
- SD
-
- stable disease
-
- PD
-
- progressive disease
-
- SPATA46
-
- spermatogenesis associated 46
-
- LMNA
-
- lamin A/C
-
- ETV6
-
- ETS translocation variant 6
Although fusion events involving neurotrophic receptor tyrosine kinase 1, 2, and 3 genes (NTRK1, NTRK2, and NTRK3, encoding TRKA/B/C respectively) were found in diverse tumor types, only 0.1%-0.3% of lung cancer patients harbor an NTRK (and mostly NTRK1) fusion as the primary oncogenic event [1]. Such low prevalence may be partially due to the limited availability of first-line assays for detecting rare fusion events [2]. Immunohistochemistry is limited by sensitivity and variable tissue background, and fluorescence in-situ hybridization falls short of elucidating functional significance such as the identity of the partner or structure of the transcript. While DNA next-generation sequencing (NGS) is suitable for mutation calling (single nucleotide variation [SNV] and insertion-or-deletion [indel]), and RNA NGS is particularly effective in detecting fusions [3], routinely performing these two assays together is labor-intensive. As a solution, we have developed a single NGS assay (PANO-Seq) for unified RNA/DNA target enrichment library preparation [4], which takes about 12 hours and $10 to prepare. Resulting libraries were pooled and sequenced using HiSeq X10 or NextSeq500/550 sequencers (Illumina, San Diego, CA, USA) with paired-end 150-cycle run setting, and raw sequencing data were analyzed using a customized data processing pipeline. Limit of detection (LoD) for mutation is 2% mutant allele frequency (MAF) using 20 ng of tissue sample input. For plasma samples, we have achieved an LoD at 0.5% MAF value.
Durable responses to tyrosine receptor kinase inhibitors (TKI) such as larotrectinib and entrectinib have been observed in a broad range of malignancies [5, 6], and the next-generation inhibitor repotrectinib exhibited promising activity against solvent-front substitution mutations [7], while inhibitors such as crizotinib initially developed for other kinases may also be effective in targeting TRK [8]. TRK-fusion protein consists of an intact oncogenic kinase domain, either constitutively activated by a 5’ partner domain or overexpressed due to a stronger partner gene promoter. Breakpoints often immediately precede the kinase domain-encoding exons, and partner genes are diverse [9]. Here, we leveraged this NGS assay to examine the status of NTRK fusions and investigated a multi-kinase inhibitor, ensartinib, for the treatment of NTRK1-positive lung cancer.
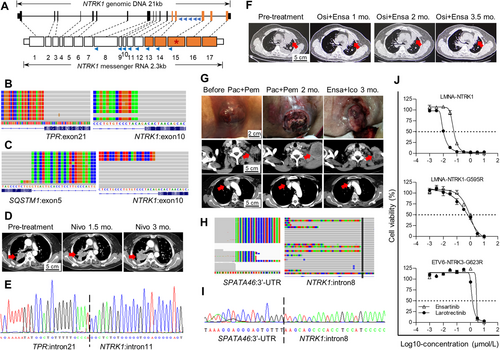
We designed a customized panel covering NTRK1 (NM_002529) exons 8-15 for unspecified upstream partners (Fig. 1A). This panel also detects mutations in epidermal growth factor receptor (EGFR), KRAS proto-oncogene (KRAS), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), and B-Raf proto-oncogene, serine/threonine kinase (BRAF), exon14-skipping of MET proto-oncogene (MET), and fusions involving anaplastic lymphoma kinase (ALK), ROS proto-oncogene 1 (ROS1), ret proto-oncogene (RET), BRAF, neuregulin 1 (NRG1), NTRK2, and NTRK3.
We assayed 1125 formalin-fixed, paraffin-embedded lung tumor samples. The selection was prioritized to samples collected within two years for assay quality purposes, without considering other criteria. This collection included 829 (74%) adenocarcinoma cases, wherein most of the genetic alterations were found (Supplementary Table S1). Read depths of a housekeeping gene, charged multivesicular body protein 2A (CHMP2A), cDNA, and gDNA were used to calculate RNA/DNA ratio, which was used to evaluate RNA quality (ratio ≥ 0.1 as pass). DNA quality was evaluated using consolidate read depth of 80% of target regions (depth > 100 reads as pass). Positive calls could also be made on some samples failed quality evaluation. Mutations in EGFR were prevalent, found in 48% (324/671) of adenocarcinoma cases after removing cases with unqualified samples and adjusting for cases harboring multiple mutations. Diverse fusion events, involving ALK, ROS1, RET, BRAF, and NRG1, and MET exon-skipping accounted for 10% (59/590) of adenocarcinomas after quality adjustment. A rare human leukocyte antigen DRB1 (HLA-DRB1)-MET fusion was also identified from this collection, demonstrating the potential robustness of this assay.
We identified only 3 cases harboring NTRK1 fusions. Among these, translocated promoter region protein (TPR)-NTRK1 fusion (Fig. 1B) and sequestosome 1 (SQSTM1)-NTRK1 fusion (Fig. 1C), in patients P1 and P2 respectively, involved previously reported partners with functional in-frame transcripts. The RING finger and WD repeat domain 2 (RFWD2)-NTRK1 fusion transcript, in patient P3, involved noncoding sequences from the partner and was likely non-functional. A frameshifted moesin (MSN)-NTRK2 transcript found in this collection of tumor samples was also considered non-functional, and no NTRK3 fusion case was detected.
Clinical records of these three patients were retrospectively examined. Patient P1 was initially admitted for stage IV adenocarcinoma, with a primary lesion in the right lung that metastasized to both lungs. This EGFR-negative patient initially received gemcitabine and cisplatin and achieved partial response (PR) for 6.7 months, then was enrolled in CHECKMATE-078, a phase III trial randomized to nivolumab [10]. Best response was stable disease (SD) on day 44, followed by progressive disease (PD) on day 90 after initiation of nivolumab treatment (Fig. 1D). A sample collected via needle biopsy showed the presence of TPR-NTRK1 fusion. Both patient P2, with stage Ia adenocarcinoma in situ in the right lung, and patient P3, with stage Ib squamous cell carcinoma in the right lower lobe, received only surgical treatments and were discharged without follow-up.
EGFR mutations represent the most common actionable mutations in Chinese lung cancer population, and resistance to targeted therapy routinely arises [11]. We hypothesized that some resistant cases could be attributed to activation of other kinases, such as TRK1, that bypass the EGFR-mediated signaling pathways, and indeed, identified two such cases (patients P4 and P5).
Patient P4, with EGFR p.L858R mutation, had a stage IV adenocarcinoma in the left lung and metastatic lesions in both lungs. This patient had undergone chemotherapy with pemetrexed and nedaplatin (SD for 2 months), followed by first-generation EGFR inhibitor erlotinib (PR for 16 months), and then the third-generation inhibitor osimertinib (PR for 17 months), until developing progressive disease (PD). Since the RNA-containing tissue specimen was no longer obtainable, a modified assay targeting NTRK1 introns 7-14 (Fig. 1A) was performed, revealing a TPR-NTRK1 fusion in the plasma cell-free DNA, validated by Sanger sequencing (Fig. 1E). In line with the bypass activation hypothesis, EGFR p.T790M, often found conferring resistance to first-generation EGFR inhibitors, was not present. Combinatorial therapy with osimertinib and ensartinib was then administered, resulting in PR in the left lung and decreased pleural effusion (Fig. 1F). The patient was assessed as SD for 2 months and PD after 5 months, declined further treatment and deceased 2 months later.
Patient P5, initially diagnosed as harboring an EGFR exon 19 deletion mutation (p.E746_A750del), presented with a stage IV adenocarcinoma in the right lung with a swollen lymph node above the left clavicle. This patient received first-line EGFR inhibitor gefitinib treatment and remained progression-free for 24 months before the emergence of p.T790M mutation and disease progression. The patient was then treated with osimertinib for 6 months until progression again. Next, the patient received, and responded poorly to, combinatorial chemo- and immunotherapy regimen with paclitaxel and pembrolizumab and developed hyper-progression in the lymph node lesion (Fig. 1G). At that point, the plasma assay (Fig. 1H) followed by Sanger sequencing (Fig. 1I) revealed a spermatogenesis associated 46 (SPATA46)-NTRK1 fusion, together with the initial p.E746_A750del mutation but not p.T790M, suggesting bypass signaling as the resistance mechanism. The patient then received ensartinib and icotinib, targeting TRK1 and EGFR respectively. Durable response was observed with marked tumor regression in both the right lung primary lesion and the lymph node metastatic lesion (Fig. 1G).
We evaluated ensartinib for TRK inhibition in Ba/F3 cell lines harboring either a wild-type lamin A/C (LMNA)-NTRK1 fusion, or a solvent-front substitution mutation, LMNA-NTRK1-G595R or ETS translocation variant 6 (ETV6)-NTRK3-G623R, conferring resistance to first-generation TRK inhibitors. The half-maximal inhibitory concentration (IC50) was 57 nmol/L for ensartinib compared to 10 nmol/L for larotrectinib for the cell line overexpressing the wild-type fusion, while viabilities of cell lines overexpressing the substitution mutations were not inhibited, indicating high target specificity (Fig. 1J).
In summary, we have demonstrated the use of a dual-template and single-workflow NGS assay as a front-line test to simultaneously detect both fusion and mutation events. We discovered NTRK1 fusions as a primary event in 0.3% of lung tumors and as a cause of acquired resistance in EGFR-positive lung cancer patients who had undergone multi-line therapies. We also presented ensartinib as a practical alternative to dedicated TRK-targeting therapies. Together, accurate detection and timely treatment could improve the clinical outcome of NTRK1-positive patients.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All patients consented to genetic testing, therapy, and registration of data. Genetic tests were performed in concordance with the ethical guidelines and reviewed by the institutional ethics committee.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
COMPETING INTERESTS
YG, WZ, and YH are employees of HeliTec Biotechnologies. LM is an employee of Betta Pharmaceutical. LC is an advisor of HeliTec Biotechnologies. The remaining authors declare no conflict of interest.
FUNDING
This work is supported by the National Natural Science Foundation of China (No. 81802276 to Z.S.).
ACKNOWLEDGMENTS
The authors thank Dr. Zongli Zheng for advice on project design and data processing, Drs. Cyril H. Benes and Maurizio Scaltriti for valuable comments on manuscript preparation, and Xuemei Zhou and Dr. Anahita Dastur for proofreading the manuscript.
AUTHORS' CONTRIBUTIONS
ZS, CX, and XP designed the study. ZS, CX, XL, and WW collected tissue samples. XP, YZ, and LW practiced patient treatment and contributed to clinical data collection. YG, WZ, and YH performed the assays and analyzed the data. LM provided the study material. LC and MC supervised the study and contributed to manuscript preparation. All authors read and approved the final manuscript.




