S100A8 promotes epithelial-mesenchymal transition and metastasis under TGF-β/USF2 axis in colorectal cancer
Abstract
Background
The transforming growth factor-β (TGF-β) pathway plays a pivotal role in inducing epithelial-mesenchymal transition (EMT), which is a key step in cancer invasion and metastasis. However, the regulatory mechanism of TGF-β in inducing EMT in colorectal cancer (CRC) has not been fully elucidated. In previous studies, it was found that S100A8 may regulate EMT. This study aimed to clarify the role of S100A8 in TGF-β-induced EMT and explore the underlying mechanism in CRC.
Methods
S100A8 and upstream transcription factor 2 (USF2) expression was detected by immunohistochemistry in 412 CRC tissues. Kaplan-Meier survival analysis was performed. In vitro, Western blot, and migration and invasion assays were performed to investigate the effects of S100A8 and USF2 on TGF-β-induced EMT. Mouse metastasis models were used to determine in vivo metastasis ability. Luciferase reporter and chromatin immunoprecipitation assay were used to explore the role of USF2 on S100A8 transcription.
Results
During TGF-β-induced EMT in CRC cells, S100A8 and the transcription factor USF2 were upregulated. S100A8 promoted cell migration and invasion and EMT. USF2 transcriptionally regulated S100A8 expression by directly binding to its promoter region. Furthermore, TGF-β enhanced the USF2/S100A8 signaling axis of CRC cells whereas extracellular S100A8 inhibited the USF2/S100A8 axis of CRC cells. S100A8 expression in tumor cells was associated with poor overall survival in CRC. USF2 expression was positively related to S100A8 expression in tumor cells but negatively related to S100A8-positive stromal cells.
Conclusions
TGF-β was found to promote EMT and metastasis through the USF2/S100A8 axis in CRC while extracellular S100A8 suppressed the USF2/S100A8 axis. USF2 was identified as an important switch on the intracellular and extracellular S100A8 feedback loop.
Abbreviations
-
- ChIP
-
- Chromatin immunoprecipitation
-
- CRC
-
- colorectal cancer
-
- EMT
-
- epithelial-mesenchymal transition
-
- IHC
-
- immunohistochemistry
-
- PBS
-
- phosphate-buffered saline
-
- PI
-
- propidium iodide
-
- SCID
-
- severe combined immunodeficient
-
- shRNA
-
- short hairpin RNA
-
- siRNA
-
- small interfering RNA
-
- S100A8
-
- S100 calcium-binding protein A8
-
- TGF-β
-
- Transforming growth factor-β
-
- USF
-
- upstream transcription factor
1 INTRODUCTION
Colorectal cancer (CRC) is the most common malignant disease of the digestive tract [1, 2]. Although its early diagnostic and treatment strategies have been improved in recent years, the morbidity and mortality of CRC have not significantly decreased. Metastasis is the most critical cause of cancer-related death [3].Efforts in studying the mechanism of CRC metastasis and searching for potential molecular targets are needed to improve the treatment and survival of CRC patients. Epithelial-mesenchymal transition (EMT) is a process in which epithelial cells lose their cell polarity and cell-cell adhesion, with reduced epithelial markers (i.e. E-cadherin), increased mesenchymal markers (i.e. Vimentin) and activated transcription factors (i.e. Snail) [4, 5]. EMT plays an important role in embryonic development, tissue healing, organ fibrosis, and tumor metastasis [6, 7]. In tumors, EMT provides malignant cells with stronger metastatic ability regulated by the complex networks between tumor cells and the tumor microenvironment [8]. Transforming growth factor-β (TGF-β) derived from tumor and stromal cell is the key regulator and major inducer of EMT [9, 10].
The microenvironment in different parts of the tumor has different regulatory mechanisms for EMT and metastasis [11]. TGF-β can induce the expression of S100 calcium-binding protein A8 (S100A8) in primary monocytes [12]. In our previous study using microdissection and gene expression microarrays, S100A8 expression in normal stroma, tumor center stroma, and tumor invasive front stroma gradually increased, suggesting that S100A8 expression could affect EMT and metastasis in CRC [13, 14]. S100A8, also known as MRP8 or calgranulin A, is a low molecular weight (10.8 kDa) calcium-binding protein and belongs to the S100 calcium-binding protein family [15]. S100A8 expression is upregulated in many tumors such as breast, pancreatic, bladder, gastric, lung, and ovarian cancers [16, 17]. However, differential S100A8 expression in tumors and the dual effects of antitumor and protumor lead to the complex relationship between S100A8 and tumors [18]. At present, most studies focus on the function of S100A8 as a secretory protein in the tumor microenvironment but its function in tumor cells is not clear. In CRC, the functions of S100A8 and its regulatory mechanism need to be clarified.
The upstream transcription factor (USF), which has two variants (USF1 and USF2) is widely expressed in eukaryotes as a transcription factor [19]. USF2 can form a dimer that binds to the E-box on the promoter of the target genes [20]. TGF-β can promote the transcriptional activity of USF2 in inflammatory response and renal fibrosis [21, 22]. USF2 can also upregulate TGF-β expression in a positive feedback and accelerate the disease process [21]. However, the relationship between USF2 and EMT has not been reported. Therefore, whether TGF-β can affect EMT by regulating the transcriptional activity of USF2 and whether USF2 can regulate S100A8 expression and promote EMT remains to be studied.
In this study, the correlation between USF2 and S100A8 in CRC and their influence on prognosis were analyzed, and the role of S100A8 in TGF-β-induced EMT was identified. Further, the role and mechanism of USF2 in the process of TGF-β regulating S100A8 were explored.
2 MATERIALS AND METHODS
2.1 Patients and tumor specimens
All 412 archival paraffin-embedded tissue blocks of CRC were collected from the First Peoples Hospital of Xiaoshan (Zhejiang, China) during 1990-2006. Information about follow-ups was provided by the Xiaoshan Centre of Disease Control, with follow-up ranging from one month to 185.29 months (median, 33 months). Two cases that died within 1 month after surgery were excluded in survival analysis. Patient and treatment data were collected from medical records. None of the patients received chemotherapy or radiotherapy before surgery. Pathological predictors were verified on the corresponding H&E slides. This study was approved by the Ethics Committee of the School of Medicine, Zhejiang University, Hangzhou, China.
2.2 Immunohistochemistry (IHC)
IHC was performed to assess the expression of S100A8 and USF2 in paraffin sections from 412 CRC cases. After deparaffinization and rehydration, paraffin-embedded tissue sections (4 μm thick) were immersed in 3% hydrogen peroxide in absolute methanol at room temperature for 15 min. Serum blocking was performed using 10% normal goat serum (Lianke Biology Technology Co., Hangzhou, Zhejiang, China) for 30 min. Primary S100A8 antibody (1:100, #DF6556, Affinity Biosciences, Changzhou, Jiangsu, China), USF2 antibody (1:300, #AF0291, Affinity Biosciences, Changzhou, Jiangsu, China), and phosphorylated Smad2 (p-Smad2, 1:100, #ZRB04953, Sigma-Aldrich, St. Louis, MO, USA) were incubated overnight at 4°C. Secondary antibody (ZB2306 or PV9000, Zhongshan Goldenbridge Biotechnology, Beijing, China) was incubated following the manufacturer's instructions. Staining was visualized with diaminobenzidine (Zhongshan Goldenbridge Biotechnology, Beijing, China). Sections were treated with hematoxylin, dehydrated, and cover-slipped. After the slides were scanned at ×20 magnifications, the cells with positively stained were counted at ×20 magnifications. The percentage of S100A8/USF2/p-Smad2 expression was quantified by determining the number of positive cells expressing S100A8/USF2/p-Smad2 among the total number of tumor cells. Samples were considered positive for S100A8/USF2/p-Smad2 when the proportion of positively stained cells was >5%. Missed data were caused by tissue falling off the section during IHC. Finally, S100A8 was detected in 407 cases, and USF2 was detected in 374 cases.
2.3 Cell culture and reagents
CRC cell lines (DLD1, HT29, HCT8, and SW480) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA), and HEK 293T cells used in transfection were from Chinese Academy of Sciences (Shanghai, China). Cells were cultured as described previously [23]. Briefly, cells were respectively cultured in RPMI 1640 (Hyclone, Logan, UT, USA) and Dulbecco's Modified Eagle's Medium (DMEM, Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS, Yeasen, Shanghai, China) and grown at 37°C under 5% CO2. All cell lines had no Mycoplasma contamination, and were identified by short tandem repeat-based methods. The CRC cells were treated with 10 ng/mL TGF-β for at least 1 week, and then collected for Western blot analysis.
2.4 Cell migration and invasion assays
Cell migration and invasion were measured as described previously [24]. Briefly, CRC cells in serum-free RPMI 1640 media were seeded into the upper chamber for migration assays (8 μm pore size; Corning Costar, Corning, NY, USA) and invasion assays with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). The lower chambers were filled with media containing 10% FBS. After several hours of incubation at 37°C, the cells that had migrated or invaded through the membrane were fixed in 4% paraformaldehyde (Sinopharm, Shanghai, China) and stained with crystal violet (Beyotime, Shanghai, China) for 10min. Migrated cells were then digested in 33% acetic acid and quantified by measuring absorbance at 570nm with a 96-well plate on a microplate reader (Bio-TEK, Winooski, VT, USA). DLD1 and SW480 cells (1 × 105) were seeded into the upper chambers and measured after 48 h. HCT8 cells (5 × 104) were seeded into the upper chambers and measured after 16 h. HT29 (2 × 105) were seeded into the upper chambers and measured after 7 days. Three independent experiments were performed.
2.5 Cell apoptosis assay
The cells were stained with Annexin V/PI apoptosis kit (70-AP101-100, MultiSciences, Hangzhou, Zhejiang, China) according to the manufacturer's protocol. Briefly, the cells were collected and washed twice with cold phosphate-buffered saline (PBS), and then resuspended in binding buffer at a concentration of 3 × 105 per tube. 10 μL propidium iodide (PI) and 5 μL Annexin V-FITC was added and was incubated for 5 min in the dark. The samples were examined by flow cytometry (DxFLEX, Beckman Coulter, Indianapolis, IN, USA). Data analysis was performed using the CytExpert for DxFLEX (Beckman Coulter, Indianapolis, IN, USA).
2.6 Protein extraction and Western blot analysis
CRC cells were lysed in radioimmunoprecipitation assay lysis buffer (Beyotime, Shanghai, China) including PMSF (Beyotime, Shanghai, China) and Protease Inhibitor Cocktail (MedChem, Monmouth Junction, NJ, USA) by sonication on ice. The lysates were centrifuged at 12000 rpm for 10 min at 4°C, and then transferred the supernatant to a new tube. Nucleoprotein was extracted using a nuclear and cytoplasmic protein extraction kit (KeyGEN, Nanjing, Jiangsu, China). The protein concentrations were determined by the BCA (Thermo Fisher, Waltham, MA, USA) method. Mixed one volume 5× loading Buffer (Fude, Hangzhou, Zhejiang, China) with four volumes of protein sample. Boiled protein samples for 3-5 min. The protein samples were stored at -80°C.
The protein expression levels were assessed by Western blot with antibodies against E-cadherin (1:1000, #3195, CST, Danvers, MA, USA), Vimentin (1:1000, #5741, CST, Danvers, MA, USA), Snail (1:500, #3879, CST, Danvers, MA, USA), p-Smad3 (1:1000, #9520, CST, Danvers, MA, USA), p-Smad2 (1:1000, #ZRB04953, Sigma-Aldrich, St. Louis, MO, USA), TGF-βR2 (1:1000, #AF5449, Affinity, Changzhou, Jiangsu, China), S100A8 (1:1000, #DF6556, Affinity, Changzhou, Jiangsu, China), USF2 (1:1000, #AF0291, Affinity, Changzhou, Jiangsu, China), GAPDH (1:5000, MultiSciences, #85-14-9523-82, Hangzhou, Zhejiang, China) and Histone (1:2000, #4499s, CST, Danvers, MA, USA). Lysate protein (20 μg) was separated on 12% SDS-polyacrylamide gels. The protein samples were subsequently transferred onto Immobilon-NC Membrane (Merck Millipore, Darmstadt, Germany) for 100 min at 100V. The membranes were blocked in TBS-Tween 20 (TBST) containing 5% low-fat milk for 1 h at room temperature and incubated in primary antibody overnight at 4°C. The blots were then incubated with IR-dye secondary antibodies (LI-COR, Lincoln, NE, USA) for 1 h at room temperature and visualized by Odyssey ® Imager (LI-COR, Lincoln, NE, USA).
2.7 Construction and transfection of overexpression/shRNA-expression plasmid
The S100A8-overexpression lentiviral plasmid (pLVX-IRES-ZsGreen vector) was constructed by Genscript Biotech Co., Ltd (Nanjing, Jiangsu, China). The S100A8-short hairpin RNA (shRNA) plasmid (pGPU6/GFP/Neo vector) was constructed by Genepharma co., ltd (Shanghai, China). The USF2-overexpression plasmid (pEnter) was provided by Vigene Biotech Co., Ltd (Jinan, Shandong, China). The USF2-shRNA plasmid (pGPU6/GFP/Neo vector) was constructed by Genepharma. The shRNA sequence is listed in Supplementary Table S1.
Cells were seeded in 6-well plates at a density of 4×105 cells per well and incubated overnight. 1 mL complete culture medium without serum was freshly added to each well 30 min before transfection. Diluted 2.5 μg of plasmid DNA into 50 μL of serum-free DMEM in tube A. Diluted 5 μL of Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA, USA) into 50 μL of serum-free DMEM in tube B. Mixed tube A and tube B followed by incubation for 5 min at room temperature and added the mixture onto the medium in each well. Removed complex-containing medium and replaced with complete serum containing medium 12 h post transfection and checked transfection efficiency 48 h after transfection.
2.8 Small interfering RNA (siRNA) transfection
All siRNAs were produced by Genepharma Co., Ltd (Shanghai, China). siRNA sequences used in this study were as follows: sense sequence 5'-CCAGGAGUUCCUCAUUCUGTT-3' and antisense sequence 5'-CAGAAUGAGGAACUCCUGGTT-3' for siS100A8; sense sequence: 5'-GCCAGUUCUACGUCAUGAUTT-3' and antisense sequence 5'-AUCAUGACGUAGAACUGGCTT-3' for siUSF2; sense sequence: 5'-UCCUCCGAACGUGUCACGUTT-3' and antisense sequence 5'-ACGUGACACGUUCGGAGAATT-3' for control (si-Ctrl). Cells were seeded in 6-well plates at a density of 2×105 cells per well and incubated overnight. The next day, cells were transfected with 50 nM S100A8 siRNA, USF2 siRNA, or non-targeting siRNA using GenMute siRNA Transfection Reagent (SignaGen, Frederick, MD, USA).After 48 h, the cells were collected for the following experiments.
2.9 Immunofluorescence assay
CRC cells plated on coverslips were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, followed by permeabilization with 0.1% Triton X-100 for 20 min. The cells were blocked with 10% bovine serum albumin in PBS for 30 min and incubated with primary antibody overnight at 4°C (S100A8 antibody, 1:400, #DF6556, Affinity Biosciences, Changzhou, Jiangsu, China; FLAG-M2 antibody, 1:200, #F1804, Sigma-Aldrich, St. Louis, MO, USA). After washing three times with PBS, the cells were incubated with Alexa Fluor 488 Donkey anti-Mouse IgG (1:300, #A-21202, Thermo Fisher, Shanghai, China) or Alexa Fluor 546 goat anti-Rabbit IgG (1:500, #A-11010, Thermo Fisher, Shanghai, China) for 1 h at room temperature. Then, the cells were counterstained with 4,6-diamino-2-phenylindole (DAPI, 1:5000, #D1306, Thermo Fisher, Shanghai, China) for 10 min before the images were captured with a confocal microscope (LSM 880, Zeiss, Oberkochen, Germany).
2.10 Luciferase reporter assay
HEK 293T cell whole-genome DNA was extracted using the TIANamp genomic DNA kit (Tiangen Biotech, Beijing, China). The predicted USF2 binding sequence on the S100A8 promoter (−657/−374 bp upstream from the transcriptional start site) was amplified. After Nhel and HindIII double-enzyme (Thermo Fisher, Waltham, MA, USA) digestion, each digested sequence was constructed on the PGL3 vector named PGL3/S100A8. PGL3/S100A8 was transfected with the pRL-TK vector and USF2 overexpression plasmid into HEK 293T cells. After 48 h, the HEK 293T cells were digested by lysis solution. The lysate was detected by LB9507 Luminometer (Berthold, Bad Wildbad, Germany) with the double-luciferase reporter gene detection kit (Beyotime, Shanghai, China).
2.11 Chromatin immunoprecipitation (ChIP)
ChIP was performed using the ChIP-IT Express kit (#102026, Active Motif, Carlsbad, CA, USA). Briefly, USF2-overexpressing HEK 293T cells were fixed with 4% formaldehyde. After fixing, the cells were scraped off and centrifuged (3500rpm) for 5 min at 4°C. Cell precipitation was resuspended in lysis solution, cleaved for 30 min at 4°C, and centrifuged (5000 rpm) for 5 min at 4°C. The precipitate was resuspended in ultrasonic lysate and ultrasonic for 20 times. After centrifugation (15000 rpm) for 10 min at 4°C, the supernatant was analyzed by PCR. The primers used for PCR were as follows: forward primer 5'-TATGGCCTGACCACCAATGC-3' and reverse primer 5'- TCTCCCTGCCAGAGTTGCTA -3' for the S100A8 promoter; forward primer 5'-GGGACCAGGAGACCATTTAGAA-3' and reverse primer 5'-TGTCATGTCAGAAATGGCGTTG-3' for control.
2.12 Animal models for pulmonary metastasis
Four-week-old male severe combined immunodeficient (SCID) mice (Slack, Shanghai, China) with uniform weight were used to generate a mouse pulmonary metastasis model. HCT8 cells were trypsinized and resuspended in PBS at a density of 3 × 104 cells/μL. Then, 100 μL of the suspension was injected into the lateral tail vein of each mouse. After 8 weeks of breeding, all the mice were euthanized. Their whole lung was collected and fixed with 4% formalin. After paraffin embedding and tissue slicing, tissue sections were stained with hematoxylin and eosin. The number of lung metastasis foci was counted under a light microscope.
2.13 Bioinformatic analysis
The correlation analysis of S100A8, USF2, and TGF-β expression was analyzed using COAD and READ Datasets from Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn). USF2 binding sites in the S100A8 promoter were predicted using the JASPAR database (http://jaspar.genereg.net/).
2.14 Statistical analysis
Comparisons between the clinicopathological variables and IHC score of protein expression were performed using the χ2 test or Fisher's exact test using SPSS 22.0 statistical software (IBM SPSS, Armonk, NY, USA). Univariate survival analyses were performed and overall survival curves were constructed using the Kaplan-Meier method with the log-rank test. Student's t-test was used in Western blot, cell apoptosis assays, and cell migration and invasion analyses. P <0.05 were considered statistically significant.
3 RESULTS
3.1 S100A8 is associated with CRC progression and poor survival
S100A8 expression was detected in 407 CRC specimens by IHC. The results showed that 82 (20.1%) samples had cytoplasmic S100A8 expression in their tumor cells (Figure 1A and B). The correlation between S100A8 expression and clinicopathological characteristics of CRC patients are summarized in Table 1. Comparing to S100A8-negative group, the patients in S100A8-positive group had more lymph node metastasis (P = 0.021), more distant metastasis (P = 0.018), higher TNM stage (P = 0.009), and worse overall survival rate (P = 0.033). Furthermore, S100A8 expression was detected in 41 lymph node metastases and 3 liver metastases (Figure 1C and D). The positive rate of S100A8 in lymph node metastases (31/41, 75.6%) was significantly higher than that in the primary tumors (82/407, 20.1%; P < 0.001).

| Characteristics | Total | S100A8 expression in tumor cells (cases [%]) | P value | ||
|---|---|---|---|---|---|
| Negative | Positive | ||||
| Age (years) | 0.964 | ||||
| <60 | 153 | 122 (37.5) | 31 (37.8) | ||
| ≥60 | 254 | 203 (62.5) | 51 (62.2) | ||
| Gender | 0.389 | ||||
| Male | 221 | 173 (53.2) | 48 (58.5) | ||
| Female | 186 | 152 (46.8) | 34 (41.5) | ||
| Lymph node metastasis | 0.021 | ||||
| Absent | 215 | 181 (55.7) | 34 (41.5) | ||
| Present | 192 | 144 (44.3) | 48 (58.5) | ||
| Distant metastasis | 0.018 | ||||
| Absent | 366 | 298 (91.7) | 68 (82.9) | ||
| Present | 41 | 27 (8.3) | 14 (17.1) | ||
| TNM stage (8th edition, AJCC) | 0.009 | ||||
| I | 78 | 71 (21.8) | 7 (8.5) | ||
| II | 123 | 99 (30.5) | 24 (29.3) | ||
| III | 165 | 128 (39.4) | 37 (45.1) | ||
| IV | 41 | 27 (8.3) | 14 (17.1) | ||
| Histological grade | 0.904 | ||||
| Low | 295 | 236 (72.6) | 59 (72.0) | ||
| High | 112 | 89 (27.4) | 23 (28.0) | ||
| Vessel infiltration | 0.751 | ||||
| Absent | 347 | 278 (85.5) | 69 (84.1) | ||
| Present | 60 | 47 (14.5) | 13 (15.9) | ||
| Overall survival status | 0.033 | ||||
| Alive | 314 | 258 (79.4) | 56 (68.3) | ||
| Dead | 93 | 67 (20.6) | 26 (31.7) | ||
Kaplan-Meier survival analysis showed that 26 (32.1%) patients in the S100A8-positive group died, whereas 67 (20.7%) patients in the S100A8-negative group died. S100A8-positive expression had the potential to predict unfavorable outcome of CRC patients (P = 0.069; Figure 1E).
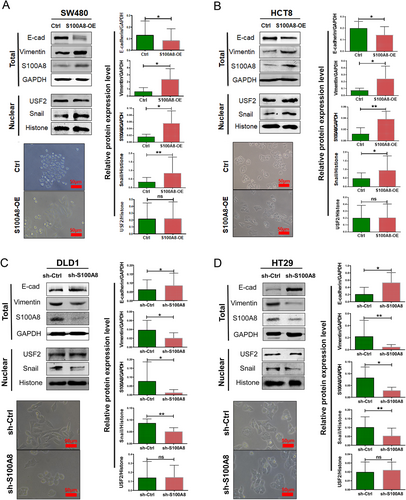
3.2 S100A8 promotes migration and invasion in CRC
S100A8 was overexpressed in CRC cell lines SW480 and HCT8 with low S100A8-expression and knocked down in DLD1 and HT29 cell lines with high S100A8-expression. Transwell assay showed that S100A8 overexpression promoted cell migration and invasion (Figure 1F and G) whereas S100A8 knockdown inhibited cell migration and invasion (Figure 1H and I). Moreover, S100A8 knockdown had no effect on apoptosis but S100A8 overexpression induced apoptosis (Supplementary Figure S1).
To further investigate the in vivo function of S100A8, S100A8-overexpressed and control HCT8 cells were injected into the lateral tail vein of SCID mice. Four mice in the S100A8 overexpression group developed lung metastases, with 7, 9, 10, and 15 metastatic foci, respectively. However, one mouse in the control group did not develop lung metastasis, and the number of metastatic foci in the other three cases was 1, 4, and 9, respectively. There was a significant difference in the number of metastases between the two groups (Figure 1J).
3.3 S100A8 induces EMT to facilitate migration and invasion in CRC
To clarify how S100A8 affects migration and invasion in CRC, cell morphology was accessed through an optical microscope and EMT markers were detected by Western blot in S100A8-overexpressed CRC cells. As shown in Figure 2A and B, S100A8-overexpressed SW480 and HCT8 cells displayed a spindle-like morphology. Western blot showed that E-cadherin was decreased, whereas Vimentin and nuclear Snail (an EMT related transcription factor) was increased in S100A8-overexpressed CRC cells. In contrast, S100A8 knocked-down DLD1 and HT29 cells displayed a rounded morphology (Figure 2C and D). Western blot showed that E-cadherin was increased, whereas Vimentin and nuclear Snail was decreased (Figure 2C and D), suggesting that S100A8 could activate EMT and thereby influence the metastasis of CRC cells.
3.4 USF2 transcriptionally regulates S100A8 expression
USF2 is a classic transcription factor that binds to the target gene promoter with the specific sequence (E-box) and enhances the transcription of downstream genes. USF2 was predicted to bind with the S100A8 promoter sequence by the JASPAR database, hinting that USF2 could transcriptionally regulate S100A8 (Supplementary Figure S2A). S100A8 overexpressed or knocked-down in CRC cells did not change the USF2 expression (Figure 2A-D).
To verify whether USF2 could combine the predicted binding sequence of S100A8 and regulate the transcription of S100A8, the PGL3/S100A8 luciferase reporter gene system was constructed and transfected into HEK 293T cells. When USF2 was overexpressed in HEK 293T cells, the activity of PGL3/S100A8 was increased (Figure 3A). Furthermore, ChIP experiments showed that USF2 directly bounded to the S100A8 promoter (AACACGTGTCC; Figure 3B). When this sequence was mutated, the luciferase reporting activity decreased significantly (Figure 3C).
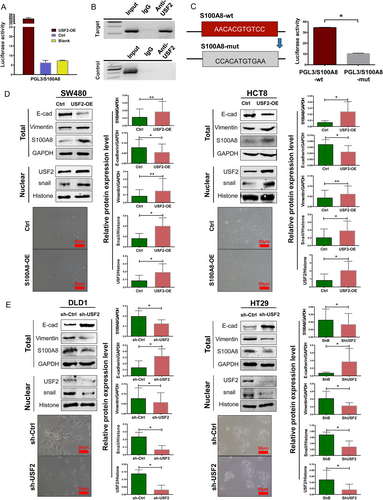
USF2 was overexpressed in SW480 and HCT8 cells and knocked down in DLD1 and HT29 cells. Western blot (Figure 3D and E) and immunofluorescence staining (Supplementary Figure S2B) showed that USF2 overexpression upregulated S100A8 expression, and USF2 knockdown downregulated S100A8 expression.
3.5 USF2 prompts EMT and predicts poor survival
After USF2 overexpression, cancer cells lost cell-cell contact and displayed EMT-like morphological features. Western blot showed that USF2 overexpression inhibited E-cadherin and upregulated Vimentin and nuclear Snail (Figure 3D). In contrast, USF2 knockdown alleviated EMT (Figure 3E). Transwell assay showed that USF2 increased cell migration and invasion (Figure 4A and B). Moreover, USF2 knockdown promoted apoptosis, but USF2 overexpression decreased apoptosis (Supplementary Figure S3).
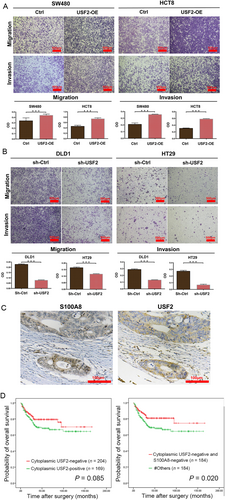
IHC staining of USF2 was performed on 374 CRC tissues (Figure 4C). USF2 was expressed in the cytoplasm or nucleus. The positive rate of cytoplasmic USF2 was 45.2% (169/374), and the positive rate of nuclear USF2 was 8.3% (31/374). Of the 72 cases of S100A8-positive tumor cells, 52 (72.2%) cases of cytoplasmic USF2 were positive. There was a significant positive correlation between cytoplasmic USF2 and S100A8 (P < 0.001, r = 0.272). However, there was no significant correlation between nuclear USF2 and S100A8 expression (P = 0.676). Cytoplasmic USF2 expression was negatively correlated with survival status (P = 0.044) but not related to the other clinicopathological parameters (Supplementary Table S2). Kaplan-Meier survival curve analysis showed that patients with negative cytoplasmic USF2 expression tended to have longer survival time (P = 0.085; Figure 4D). Furthermore, patients with both negative cytoplasmic USF2 and S100A8 expression had better survival than the others (P = 0.020; Figure 4D).
3.6 USF2 switches the intracellular and extracellular functions of S100A8
Our previous study showed that S100A8 was also expressed in the tumor stroma, especially on neutrophils. Additionally, a strong infiltration of S100A8-positive stromal cells in the tumor invasive front was associated with favorable outcome in CRC patients [14]. Extracellular S100A8 inhibited EMT and induced apoptosis of tumor cells in vitro [14]. In contrast, this study showed that S100A8 expressed in tumor cells decreased the overall survival time for CRC. In vitro studies revealed that S100A8 in tumor cells promoted EMT and metastasis. Therefore, we further explored the effect of USF2 on the intracellular and extracellular functions of S100A8.
After recombinant human S100A8 (2 μg/mL) was added into the cell culture medium, the intracellular USF2/S100A8 expression was inhibited (Figure 5A) and the EMT was restrained but similar inhibition was not observed in USF2-overexpressed SW480 cells (Figure 5B).
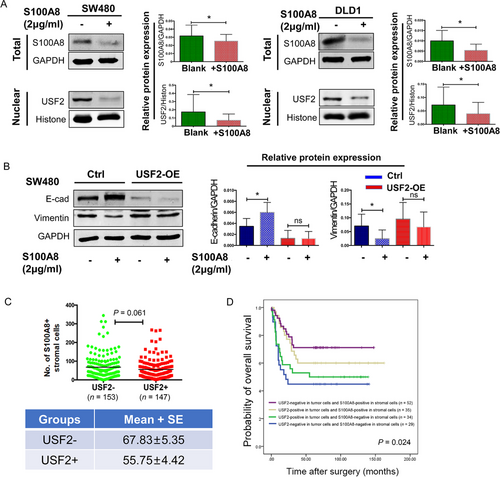
Although IHC results showed that USF2 expression in tumor cells was positively related to S100A8 in the tumor cells, the positive group of USF2 in tumor cells had a lower density of S100A8 stromal cells (Figure 5C). Patients with negative USF2 expression and more S100A8-positive stromal cells had the best survival (Figure 5D).
3.7 TGF-β induces EMT by up-regulating the USF2/S100A8 axis
TGF-β is a classic inducer of EMT [10]. To determine whether the USF2/S100A8 axis plays a role in TGF-β-induced EMT, data mining was performed using the GEPIA web server. The correlation analysis indicated that TGF-β expression was positively related to the expression of USF2 and S100A8 (Supplementary Figure S4A and B). p-Smad2 is a key intracellular signal molecule and transcription factor for TGF-b signaling. [25]. p-Smad2, USF2, and S100A8 expression also was detected by Western blot in 4 CRC cell lines and 10 CRC specimens. Similarly, the expression of p-Smad2 detected by Western blot was positively related to the expression of USF2 and S100A8 in CRC cell lines and CRC specimens (Supplementary Figure S4C and D). The correlations among p-Smad2, USF2, and S100A8 were analyzed in 100 CRC specimens by IHC. The results showed that nuclear p-Smad2 expression was positively correlated with cytoplasmic USF2 and S100A8 expression (Supplementary Table S3 and Figure S4E-G).
To verify the correlation between S100A8 and TGF-β, SW480 and DLD1 cells were treated with TGF-β (10 ng/mL) for 1 week. As shown in Figure 6A and B, S100A8 was increased in TGF-β-treated cells, compared to the control cells. As TGF-βR2 and p-Smad3 were both upregulated, a decrease in the epithelial marker E-cadherin and an increase in the mesenchymal marker Vimentin and transcription factor Snail was observed, indicating that TGF-β successfully induced EMT in CRC cells. TGF-β promoted the migration and invasion of control cells after treatment. However, TGF-β did not restore cell migration and invasion in DLD1 and HT29 cells with S100A8 knockdown (Figure 6C and D). These results indicated that S100A8 was involved in TGF-β-induced EMT.
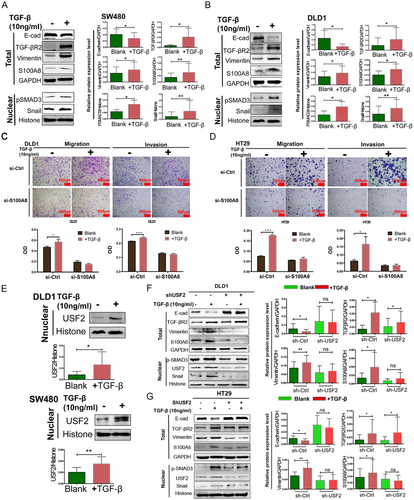
As shown in Figure 6E, TGF-β not only upregulated S100A8 expression but also increased nuclear USF2 expression in DLD1 and SW480 cells. Compared to the control cells, TGF-β neither enhanced cell migration or invasion (Supplementary Figure S5) nor induced EMT in USF2 knocked-down DLD1 and HT29 cells (Figure 6F and G). Intriguingly, S100A8, and USF2 also upregulated nuclear p-Smad2 expression in CRC cell lines (Supplementary Figure S6A and B). Overall, the results mentioned above revealed that intracellular USF2/S100A8 was the target of TGF-β on EMT and metastasis, whereas extracellular S100A8 suppressed the USF2/S100A8 axis.
4 DISCUSSION
This study demonstrated that intracellular and extracellular S100A8 had different effects on CRC prognosis, and were related to the promotion and inhibition of EMT, respectively. Extracellular S100A8 inhibited tumor metastasis and improved prognosis by promoting apoptosis and inhibiting EMT [14]. Here, S100A8 expression was increased in EMT induced by TGF-β. The presence of S100A8 in tumor cells promoted metastasis by inducing EMT, which was not conducive to patients’ prognosis. USF2 regulated S100A8 expression and played an important role in the S100A8 feedback loop.
Tumor cells live in a specific microenvironment called tumor microenvironment. The tumor microenvironment is closely related to the occurrence, development, and prognosis of tumors [9]. It is an irreplaceable factor in EMT-involved tumor metastasis [26]. Because of the heterogeneity of tumor cells and the microenvironments’ diversity, the separation of tumor cells from the surrounding environment is essential in the study of the interaction between the tumor and the microenvironment. A series of differentially expressed genes, such as tumor center cells and tumor buds, have been discovered in different tumor cells and their stromal cells in different parts of the tumor. These genes are involved in cell adhesion, tumor immunity, EMT, and many other pathways, which play important roles in tumor metastasis [13]. S100A8 is one of them and as its expression gradually increases in stromal cells around normal epithelial cells, tumor center cells, and tumor buds, this suggests that S100A8 might affect EMT and the metastasis of CRC cells.
S100A8 has been shown to have contradictory roles in tumors [18]. In CRC, higher S100A8 expression was related to lower histological differentiation, higher duke' s stage, and positive lymph node metastasis [27]. However, in breast cancer, intracellular S100A8 was found to inhibit EMT and cell motility by inducing E-cadherin expression [28]. Extracellular S100A8/A9 can trigger EMT through the MCAM/ETV4/ZEB1 axis [29]. S100A8/A9 also can induce actin polymerization to regulate the cytoskeleton rearrangement [30]. However, a recent study found that S100A8 expression could be associated with improve prognosis in esophageal cancer patients whereas S100A9 or S100A8/S100A9 dimer was not associated with prognosis [31]. S100A8 is expressed in both stromal cells and tumor cells in several cancers [16]. Previously, we found that high densities of S100A8 in the stromal cells improved the prognosis of CRC patients, and involved in inhibiting tumor metastasis and EMT [14]. Moreover, in this present study (data not shown), S100A8 expression in tumor cells was negatively correlated with macrophages’ density in the tumor invasive front. Herein, positive S100A8 expression in tumor cells was unfavorable for the outcome of CRC patients. In vivo experiments in mice further confirmed that S100A8 overexpression promoted lung metastasis of tumor cells.
Originally, S100A8 was found to be produced in macrophages against bacterial attack. Once MYD88-dependent and MYD88-independent pathways are activated, S100A8 expression increases and was assisted by transcription factors such as C/EBPs [32-34]. S100A8 is a nuclear factor-κB target gene but needs the synergistic function of other transcriptional regulators [35]. Although YAP did not bind on S100A8 promoter sites, the activated Hippo pathway upregulated S100A8 expression in squamous cell carcinoma [36]. The ubiquitin ligase RNF5 interacted with S100A8 and promoted its degradation [37]. The regulatory mechanism of S100A8 in cancer cells is far from clear. S100A8 bounded to cytoskeletal proteins and participated in the cytoskeletal rearrangement, thus increasing the mobility of white blood cells and the migration of phagocytes across the endothelium [38]. Studies have also shown that the S100A8/S100A9 complex helped CD11b-CD18 on the surface of white blood cells could bind to intercellular adhesion molecule-1 of vascular endothelial cells and destroy the integrity of endothelial cells [39]. Thus, such unfavorable results regarding S100A8 in tumor cells, and its influence on the cell movement was investigated. We observed that cell migration and invasion significantly increased after S100A8 overexpression in CRC cells but were significantly decreased after S100A8 knockdown. This indicated that intracellular S100A8 promoted the movement of CRC cells and helped metastasis.
Many signaling factors, such as TGF-β, hepatocyte growth factor, and epidermal growth factor, derived from tumor stroma and tumor cells, could activate EMT-related transcription factors such as Snail and Slug, in tumor cells, then affect EMT markers and cell functions [5, 6, 8]. TGF-β in the tumor microenvironment can be secreted by tumor-related fibroblasts, platelets, and tumor cells, which is very important in EMT activation [4, 5, 10]. TGF-β has an anti-cancer effect in normal tissues, but it will lose its anticancer effect in the process of malignant transformation and thereby promote cancer [40, 41]. The transformation of TGF-β from antitumor to protumor is similar to the dual characteristics of S100A8. Our results also showed that TGF-β induced EMT in CRC cells and increased S100A8 expression. Cell migration and invasion were also enhanced by TGF-β. However, TGF-β did not restore cell migration and invasion in S100A8 knocked-down cells, indicating that TGF-β regulated EMT through S100A8.
The influence of the TGF-β/S100A8 axis on EMT and metastasis might be affected by some transcription factors. The transcription factor USF2 was involved in TGF-β/S100A8-induced EMT because USF2 not only participated in the renal interstitial fibrosis by TGF-β [21] but was also predicted to bind the E-box on the S100A8 promoter. USF2 belongs to the transcription factor of the basic helix-loop-helix-leucine zipper family [42] which is related to malignancy. USF2 inhibited tumors in prostate, breast, and oral cancers [43-45] but promoted tumors in lung cancer [46]. Little is known about the mechanism of USF2 activation or inhibition in the process of CRC. Cytoplasmic USF2 was found to have the potential to shorten survival time. However, nuclear USF2 had no relation to survival, probably due to little nuclear staining frequency. These results revealed that cytoplasmic USF2 also had functions in CRC. ChIP experiments confirmed that USF2 directly bounded to the S100A8 promoter. USF2 also enhanced cell migration and invasion, induced EMT, and upregulated S100A8. Our work highlights the role of USF2/S100A8 in promoting metastasis and uncovers the direct regulation of S100A8 by USF2, hinting a potential target for inhibiting cancer progression.
IHC results showed that cytoplasmic USF2 was positively related to S100A8 in tumor cells but negatively related to S100A8 in stromal cells. Whether USF2 plays a switching role in the dual functions of S100A8 was investigated. It was confirmed that TGF-β increased both S100A8 and USF2 expression. Furthermore, TGF-β restored neither migration and invasion nor S100A8 expression and EMT in USF2 knocked-down cells. Thus, TGF-β enhanced the USF2/S100A8 axis on EMT and metastasis. In contrast, exogenous S100A8 protein inhibited EMT and USF2 and S100A8 expression, but this inhibition did not occur in USF2 overexpressed cells.
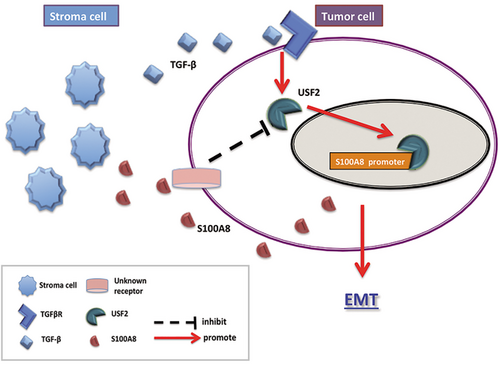
A negative feedback model was concluded based on the above results: TGF-β in the tumor stroma regulated S100A8 expression through USF2 to promote EMT. If there were redundant S100A8 secreted by tumor or stroma cells in the tumor microenvironment, it would inhibit the USF2/S100A8 pathway of tumor cells, thus inhibiting EMT (Figure 7). This negative feedback regulation explained why S100A8 plays different roles in the tumor microenvironment and tumor cells.
5 CONCLUSIONS
S100A8 induces EMT and promotes metastasis in CRC cells. Its high expression was associated with unfavorable prognosis in CRC. S100A8 was regulated by the TGF-β/USF2 axis whereas excessive extracellular S100A8 inhibited the TGF-β/USF2 axis.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Ethics Committee of the School of Medicine, Zhejiang University (Hangzhou, Zhejiang, China). Each participant signed informed consent before participating in this study. Animal care and experimentation were conducted in accordance with the guidelines of the Institutional Committee for the Ethics of Animal Care and Treatment at Zhejiang University.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study are available from the corresponding author upon reasonable request.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This work was supported by the grants of the National Natural Science Foundation of China (81772570), the Open Projects of State Key Laboratory of Molecular Oncology (SKL-KF-2019-17), and the Program of Introducing Talents of Discipline to Universities (B13026).
AUTHORS' CONTRIBUTIONS
S.L. performed mostly experiments and wrote the manuscript. J.Z. and S.Q. performed in vitro experiments and statistical analysis. X.W. performed animal experiments. L.S. performed immunohistochemical staining. T.L., Y.J., and W.L. performed immunohistochemistry. L.S. provided experimental materials support. M.L. and F.X. designed the experiments and wrote the paper. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
Thanks for the technical support by the Core Facilities, Zhejiang University School of Medicine.




