Cellular heterogeneity and inflammatory profiles in gliomas: Single-cell transcriptomic insights
Graphical Abstract
This study investigates the transcriptional profiles of gliomas across different grades (WHO II-IV) and clinical states (primary vs. recurrent). Utilizing RNA-seq data from public databases (e.g., GEO), we analyzed low-grade gliomas and high-grade gliomas, including oligodendrogliomas, glioblastomas, and other glioma subtypes. Key analyses encompassed differential gene expression, glioma subpopulation characterization (e.g., glioma-associated microglia/macrophages), regulatory network construction (WGCNA and transcription factor activity), and cell state analysis comparing primary and recurrent gliomas. Our findings reveal distinct transcriptional signatures and identify potential biomarkers associated with glioma progression and recurrence.
Dear Editor,
Gliomas are challenging brain tumors known for their aggressive nature and resistance to treatment. They exhibit substantial heterogeneity, both between different grades and within individual tumors at different stages, further complicating their clinical management. Gliomas are classified into different grades, ranging from low-grade (I and II) to high-grade (III and IV) tumors,1 based on their histological features and prognosis. Low-grade gliomas (LGGs) progress slowly and have a relatively better prognosis, while high-grade gliomas (HGGs), such as glioblastoma multiforme (GBM), are associated with a poor prognosis and limited treatment options.2 Hence, understanding the cellular and molecular heterogeneity within gliomas is crucial for developing improved diagnostic and therapeutic strategies. Single-cell transcriptomics is a cutting-edge technology that enables the profiling of gene expression patterns in individual cells. It offers a powerful tool to investigate the cellular heterogeneity within gliomas, allowing researchers to identify and characterize different cell types, their states, and their interactions within the tumor microenvironment.
In this research letter, we present a concise report of a novel discovery using single-cell transcriptomic data to explore the cellular landscape of gliomas (Figure 1), with a specific focus on different grades, states (primary and recurrent), and associated features of brain gliomas. We particularly emphasize LGGs and HGGs, as well as glioma-associated microglia/macrophages (GAMs). By analyzing 10 glioma samples (Supporting Information S1: Table S1 and Figures S1–S3), totaling 21,071 cells, we identified seven major cell types within gliomas, including oligodendrocytes, astrocytes, neural progenitor cells, glioma-associated small glial cells and macrophages, endothelial cells, and mesenchymal cells (Figure 1A,B). This findings highlight the complex cellular landscape of gliomas and reveal distinct differences in cell proportions and communication intensity between LGGs and HGGs (Figure 1C–F and Supporting Information S1: Figure S4). Notably, we identified substantial variations in the cell composition across different grades of gliomas. Specifically, within in HGGs, a noteworthy increase was observed in the proportion of GAMs, whereas the proportion of neural progenitor cells and oligodendrocytes decreased in comparison to LGGs (Figure 1C). This observation underscores the significance of alterations in cell types during the transformation of glioma. Furthermore, we conducted a comprehensive analysis to explore the distinctions in gene expression profiles between LGG and HGG (Figure 1F,G and Supporting Information S1: Figure S5). Results revealed a considerable proportion of differentially expressed genes in LGG gliomas to be associated with mitochondrial-related genes (MT genes), whereas in HGG gliomas, the differentially gene expression primarily featured ribosome-related genes (RP genes) (Figure 1G).
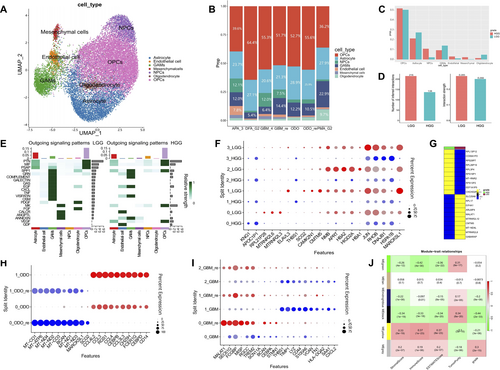
Cell type annotation, composition, and transcriptional heterogeneity in gliomas. (A) Cell type annotation of glioma samples. (B) Bar plot showing the proportional distribution of different cell types across various glioma subtypes. (C) Proportional distribution of cell types in LGG and HGG. (D) Visualization of differential gene expression in GAMs subpopulations, comparing LGG and HGG groups. (E) Cell-cell communication interactions and interaction strength in LGG and HGG. (F) Different outgoing signaling patterns in LGG and HGG groups. (G) Heatmap of differential gene expression in GAMs, comparing LGG and HGG groups. (H and I) Comparison of differential gene expression in GAMs subpopulations between primary and recurrent ODO (H) and GBM (I) samples. (J) WGCNA showing gene expression modules associated with specific immune characteristics. GAMs, glioma-associated microglia/macrophages; GBM, glioblastoma multiforme; HGG, high-grade gliomas; LGG, low-grade gliomas; WGCNA, weighted gene co-expression network analysis.
The heterogeneity of gliomas extend beyond inter-grade differences to include substantial intra-tumoral diversity within both primary and recurrent manifestations of the disease. The presence of the blood-brain barrier poses a unique challenge to the infiltration of systemic immune constituents, conferring a critical importance to GAMs within the neuro-immunological niche.3 In our investigation, we employed unbiased clustering techniques to categorize GAMs into four subpopulations, enabling the identification of distinct gene expression profiles and suggesting potential functional variations within these groups (Figure 1F). Furthermore, we conducted an in-depth analysis of GAM subpopulations across primary gliomas, recurrent gliomas, and specifically within grade IV gliomas and their recurrent counterparts (Figure 1H,I). This analysis has provided insights into their diversity and potential functional alterations of GAMs associated with disease progression. In both ODO and GBM, each GAMs subpopulations exhibited distinct gene expression signatures and distinct differentiation pathways. Importantly, when comparing primary and recurrent gliomas, we observed significant differences in the proportions of specific GAM subpopulations, as well as their differentiation trajectories (Supporting Information S1: Figure S6), highlighting the adaptive nature of GAMs in response to tumor recurrence.
Advances in molecular profiling techniques, such as genomic sequencing and transcriptomics, have provided insights into the molecular subtypes, genetic alterations, and signaling pathways associated with different glioma subpopulations.4, 5 Additionally, by utilizing weighted gene co-expression network analysis, we pinpointed two key gene modules that are intimately associated with immune infiltration and the characteristics of the tumor microenvironment in gliomas (Figure 1J). The first module is enriched with genes crucial for axon development and neuronal regulation, indicating a potential role in neural pathway modulation within the tumor milieu (Supporting Information S1: Figure S7A). The second module encompasses genes that are integral to the immune response, suggesting a significant impact on immune cell activity and interactions within the glioma microenvironment (Supporting Information S1: Figure S7B).
Overall, our research provides a granular view of the cellular and molecular landscape of gliomas through single-cell transcriptomic data, offering a window into the cellular heterogeneity and inflammatory responses that characterize these tumors across varying grades and states. The identified bright spots in cell communication, gene modules, and GAMs subpopulations could shape the development of targeted therapeutic strategies for glioma management.
AUTHOR CONTRIBUTIONS
Jiang Li: Formal analysis; writing—original draft; conceptualization; methodology. Liwei Qin: Visualization; investigation. Kailong Xu: Formal analysis; visualization. Jie Liu: Formal analysis; data curation. Anren Xu: Validation; visualization. Yunlong Qu: Validation; visualization. XiaoLu Fu: Data curation. Peng Wang: Funding acquisition. Yang Wang: Funding acquisition; writing—review & editing; supervision; project administration.
ACKNOWLEDGMENTS
This work was supported by the Hubei University Fund (202110703000002), Open Funding Project of the State Key Laboratory of Biocatalysis and Enzyme Engineering (SKLBEE20220021), and Knowledge Innovation Program of Wuhan-Shuguang Project (2022020801020325).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ETHICS STATEMENT
The data in this study are available from NCBI (https://www.ncbi.nlm.nih.gov/), therefore the ethics approval is not needed in this study.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in GEO database at https://www.ncbi.nlm.nih.gov/geo/, reference number 31554641; 31554641; 27806376. These data were derived from the following resources available in the public domain: GEO database, https://pubmed.ncbi.nlm.nih.gov/31554641/—GEO database, https://pubmed.ncbi.nlm.nih.gov/27806376/.