Circadian rhythms and mood: Opportunities for multi-level analyses in genomics and neuroscience
Circadian rhythm dysregulation in mood disorders provides clues to the brain's organizing principles, and a touchstone for genomics and neuroscience
Abstract
In the healthy state, both circadian rhythm and mood are stable against perturbations, yet they are capable of adjusting to altered internal cues or ongoing changes in external conditions. The dual demands of stability and flexibility are met by the collective properties of complex neural networks. Disruption of this balance underlies both circadian rhythm abnormality and mood disorders. However, we do not fully understand the network properties that govern the crosstalk between the circadian system and mood regulation. This puzzle reflects a challenge at the center of neurobiology, and its solution requires the successful integration of existing data across all levels of neural organization, from molecules, cells, circuits, network dynamics, to integrated mental function. This essay discusses several open questions confronting the cross-level synthesis, and proposes that circadian regulation, and its role in mood, stands as a uniquely tractable system to study the causal mechanisms of neural adaptation.
Also watch the Video Abstract.
Editor's suggested further reading in BioEssays Major depressive disorder: A loss of circadian synchrony? Abstract.
Abbreviations
-
- BD
-
- bipolar disorder
-
- GWAS
-
- genome-wide association studies
-
- MDD
-
- major depressive disorder
-
- SCN
-
- suprachiasmatic nucleus
Introduction
A hallmark feature of mood disorders is disrupted circadian/sleep patterns 1, a comorbidity well recognized at the behavioral level. However, the causal relationship between the two conditions (i.e. which comes first?) is not fully understood. Our recent study of postmortem brain tissues identified 24-hour cyclic patterns of gene expression in healthy donors, and found that such patterns were disrupted in patients of major depressive disorder (MDD) 2 (also see 3). This result uncovered the link between circadian rhythm and mood at the level of gene regulation, and drew attention to many bigger questions: How do sleep disruption and other circadian abnormalities contribute to mood disorders in general? What is the role of neural and physiological rhythms in mental health? And how can genomic datasets and systems biology help us answer these questions?
Although exciting progress has been made in clarifying the biochemical and cellular mechanisms of brain functions, neural network properties remain difficult to study, and will be the focus of this essay. I will briefly review the current state of knowledge of circadian system and mood regulation, highlighting network properties and system dynamics as key mediators of neural plasticity, which underlies the circadian rhythm-mood connection. I will provide a quick tour of relevant gene expression and genetic studies, summarizing their progress and limitations. I will describe opportunities to achieve at least modest successes in bridging the gene-level, cell-level, and circuit-level research, and discuss open questions that represent major hurdles facing the synthesis of psychiatric genetics and systems neuroscience. Throughout the essay, I will make the case that circadian regulation is an ideal system for the initial tackling of these challenges.
Plasticity, hierarchical networks, and dynamics are essential features of circadian regulation
Physiological and behavioral circadian rhythms are maintained by a biological timekeeping capability that has evolved in most of life on earth. In mammals, these rhythms are controlled by the master oscillator, which consists of a group of rhythmic cells located in the suprachiasmatic nucleus (SCN) of the hypothalamus 4. SCN rhythms are entrained by photic input from the eye. Local oscillators in other brain regions and peripheral tissues receive input from the SCN and integrate this signal with other internal cues, such as body temperature and hormone levels. Compared to other brain functions such as emotion and cognition, circadian rhythm regulation is a particularly attractive model to study. Its input can be readily controlled by adjusting the timing and quality of ambient light. Its readout is measurable, for example, as daily oscillations of hormone levels or transcriptomic patterns in a given tissue. Because circadian regulation is universal across most life forms it is relatively easy to find animal models that are generalizable to humans. For example, key genes involved in intrinsic cellular rhythms in humans (i.e. “clock genes”) have homologs in fruit flies and rodents; both are well-developed systems for genetic and functional dissection. The cellular and circuit-level organization of mammalian circadian regulation has been extensively reviewed 5. Of primary interest to this essay are the higher-level features of the circadian system – functional plasticity, network properties, and dynamics – and their relevance to other integrated mental functions.
Functional plasticity: A good clock is reliable and adjustable
Circadian regulation is a quintessential example of functional plasticity, as it demonstrates the balance between consistency and flexibility. The baseline rhythms, particularly in the SCN, are robust to genetic and environmental perturbations 6, 7. However, these rhythms can adapt to major changes in the operating environment, including seasonal changes of day length, and, in the case of modern humans and experimental animals, sudden changes in daily light schedule 5, 8. Evolution has enabled the circadian system to respond to gradual seasonal changes. However, rapid phase resetting as demanded by air travel or shift work is an evolutionarily recent, novel stressor; and the mammalian circadian system is capable of responding to such acute demands. Even so, while most people can make the transition to a new schedule, frequent shifts can lead to long-term health consequences, such as increased risk of cardiovascular and immune dysfunction.
Network properties: No clock works alone in the circadian system
In the SCN, rhythms are maintained as both single-cell and ensemble properties. Each SCN neuron generates intrinsic rhythmicity through interlocking feedback loops involving a set of “core clock genes” and their protein products. Cell-autonomous rhythms are synchronized at the level of cell populations. The SCN exerts top-down control over other brain regions and peripheral oscillators while receiving feedback from them. Thus, the circadian control system is a hierarchical network, involving “elementary clocks” in individual cells, “ensemble clocks” in cell populations, coupling between central and peripheral oscillators, and interactions with external input 9 (Fig. 1). As an analogy, different oscillators behave like sections in an orchestra – within each section every member (the cell) is a self-sufficient musician, and every musician plays in ensemble by listening to the others and following a rhythmic input (the daily cycle of environmental light). Each section has its distinct role, but harmonizes with the other sections via a precise phase relationship. There exists a standard temporal protocol, but it is possible to vary it or to reset the phase relationship. Healthy operation of the system relies on both central control and local autonomy, and the right level of plasticity. Compared to isolated cells, ensemble rhythms of the network are more resistant to genetic or environmental perturbations 6, 7, 10, serving as a reminder that circuit-level properties are central to any effort to understand brain function.
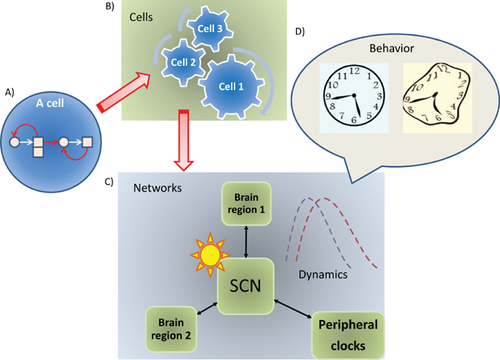
Neural dynamics: The circadian system provides an ideal model to study temporal organization
In the SCN, adaptation to seasonal changes of day length or temperature is achieved not by changed oscillating properties of individual neurons but by the altered phase relationship among them 11, 12. Circadian regulation thus provides an ideal model to study the role of temporal organization. The master-subordinate relationship between the SCN and other oscillators is known; so is the core set of 10–20 genes and how they interact with each other to maintain cell-autonomous rhythms. The next several hundred genes in specific tissues have begun to be recognized in genomic studies. There is a wealth of existing data on the wiring diagram and connection parameters for both intra- and intercellular interactions 13. The dynamic behavior of circadian oscillators has been extensively modeled (e.g. 14), allowing quantitative understanding of coupling strength, phase distribution, and their consequences 15. In the example of recovery from jet lag, one can ask some modestly ambitious questions as a starting point for understanding mechanisms of resynchronization and stability: Since the dorsal and ventral regions of the SCN show different response speeds to light schedule changes 16, how do the two circuits regain phase cohesion? How does the tissue ensemble transition from one stable state to another? Can the mechanisms of phase resetting teach us something about neural regulation in other systems?
Relevance to other systems: Everything happens in its own time, for a reason, and together
Adaptation in other mental functions, including mood, is also best understood in terms of network properties, including intrinsic dynamics, responses to stress, and alternative (meta)stable states. This essay is partly inspired by the premise that the manner in which the brain manages the synchrony and asynchrony of neuronal populations is crucial to its normal operation. Circadian rhythm regulation is not the only example where temporal patterns in neural circuits – rather than constitutional features such as static levels of neurotransmitters – encode the function of the system and mark the divide between health and disease 17, 18. In animal models, short-term dynamic modulation of synaptic plasticity is correlated with behavior in a working memory task 19. Some cardinal symptoms of depression and schizophrenia are linked to aberrant timing in information processing 20. These results strongly suggest that inter-circuit coupling and timing could be a universal mechanism for network-level function, and that impaired temporal organization could manifest as behavioral deficits 21. Further, since maladaptation may arise from an inability to regain proper synchrony, this network-level phenotype can be a target for therapy; for example, clinical data show that social rhythm stabilization can improve recovery from bipolar depression 22, 23.
Circadian rhythm and mood regulation share a similar temporal scale. The adjustment to jet lag takes many days to harmonize across oscillators, despite the fact that ventral SCN itself is entrained quickly 16. The characteristic tempo of this form of inter-circuit plasticity, spanning days to weeks, is reminiscent of the mood cycling time in bipolar disorders (BD) and the therapeutic delay observed for classic antidepressants, which take 2–4 weeks to work fully (in the case that they are effective for that individual). Could circadian phase flexibility share a common mechanism with mental resilience 24? Do patients with depression exhibit different patterns of synchronicity between oscillators compared to healthy subjects, reflecting too much or too little plasticity? Can we discover causal network features for a brain disease as opposed to causal molecular or cellular features?
Circadian system and mood disorders are linked at the behavioral and molecular levels
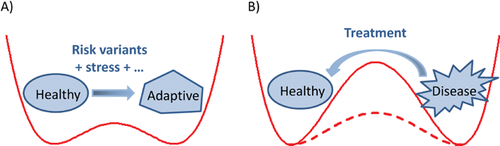
Mood disorders such as severe depression are one of the central problems in neurobiology and psychiatry. Currently, studies of mood disorder pathophysiology have implicated a staggering array of biological processes (for a review, see 25). Of particular interest to this discussion, MDD and BD engage neuroplasticity at multiple levels. At the behavioral level, the depressed state can be induced by external stress factors as an adaptive response (i.e. beneficial to the subject under the circumstance). After the initial response, many people recover to a normal state, demonstrating resilience. Yet in others, depression becomes a stable state, even resistant to treatment (Fig. 2). The exact mechanism by which the brain enters and locks into a depressed state, even when it is no longer beneficial, remains elusive. As such, proper mood regulation requires a balance between stability and plasticity, resembling the dual demand observed in circadian regulation.
Circadian abnormality can be both a causal factor and consequence of psychiatric disorders. For some people, troubled sleep adds to their stress level and provokes the onset or worsening of depression; while for others sleep disruption may be secondary to depression and/or receiving medical treatment. Clock dysfunction is closely intertwined with mood disorders in animal models. Mice carrying a dominant-negative mutation of the Clock gene show sleep disturbances, but also manic-like behavior, which can be reversed by lithium, a common mood-stabilizing drug 26. Such observations confirm a strong genetic link between circadian clock and mood regulation, although they do not resolve the causal relationship between the two in humans.
Past studies have uncovered numerous mechanisms to explain the connection between circadian rhythm disruption and mood disorders (reviewed in 27). These two functions are linked by monoamine signaling 26, 28 and immune function, as mediated by the proinflammatory cytokines 29 (but also see 30), the hypothalamic-pituitary-adrenal axis and glucocorticoids, metabolic factors such as peptides regulating eating behavior, general mitochondrial function, and neurogenesis 31, 32. Much of this impressive literature is populated by molecular and cellular findings. However, additional studies are needed to sort out the relative importance of the diverse lineup of molecular/cellular factors. In which brain circuits do they act? Under which environmental conditions? At which stage of brain development? How do they influence network dynamics and behavioral output?
What can be learned from gene expression studies?
In recent years, the challenge of knowledge integration in studies of complex traits was amplified by the influx of larger-scale data, particularly from gene expression and genetic analyses. Gene expression profiling belongs to the new discipline of Functional Genomics, which applies high-throughput methods to study the function, regulation, and interaction of all measurable molecular components of a cell or tissue system. While these components include DNA, RNA, proteins, and metabolites, here I will focus on transcriptomic changes that accompany circadian cycling.
In non-human animals, circadian cycles of gene expression have been demonstrated in the blood, brain, liver, kidney, skeletal muscle, and heart 33-36. Parallel studies in humans have been limited to easily accessible tissues such as the oral mucosa 37, skin biopsies 38, hair follicle cells 39, and cultured cell lines 40, 41. In a recent study 2, we extended whole-transcriptome analyses to the human brain, using postmortem tissues obtained from six cortical and limbic regions (not including the SCN). Our results show that circadian patterns do exist in the transcriptome of healthy donors, and are in-phase across the six regions analyzed. The patterns involve several hundred transcripts, led by the best-known clock genes such as BMAL1. The data suggest that gene expression rhythms must be stable, as the cyclicity was detected over 55 brain donors, each representing a single time point in the overall 24-hour cyclic patterns. The donors experienced different acute medical conditions at death, thus our findings re-affirm the robust and slow-changing nature of the circadian rhythm: Even though some genes respond rapidly to external factors, even on a time-scale of less than one hour 42, 43, the daily cycling of known clock genes is impervious to sudden changes in environmental cues outside of the SCN.
Our study still left many questions unanswered. First, the medical records of the study participants did not describe the quality, duration, or phase of sleep at their final days of life, precluding an answer to the what-comes-first question. Second, as a general limitation in bulk tissue analysis, studies of large brain regions can only report the average profile of all cells in the population. Future studies of the brain transcriptome need to take spatial heterogeneity into account, and must address other prominent sources of biological noise such as medication history and agonal conditions 44.
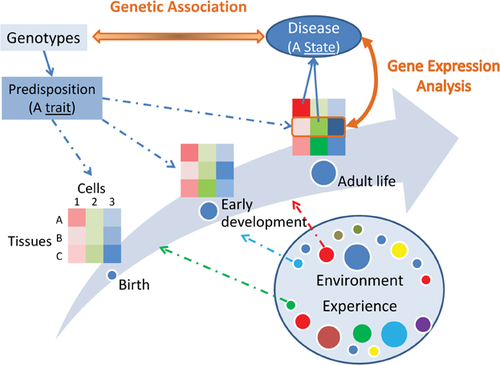
Despite these limitations, gene expression analysis remains a powerful tool for reading out the cellular and biochemical disturbances in specific brain regions – assuming that such signatures are observable at the spatial resolution attempted, and remain at the time of tissue collection. This approach provides a useful complement to the classic genetic methods because causal genetic factors only provide the innate proclivity of a trait, a yet-to-be-realized susceptibility. In contrast, gene expression profiles of the mature tissue capture the present state, and may reveal proximal causal mechanisms (Fig. 3). If the varied assortment of contributing factors, both genetic and environmental, converges on a smaller number of functional states, analysis of the transcriptomic correlates of phenotypic outcome can be more powerful and more informative than simply trying to detect the genetic correlates.
Genetic studies of circadian rhythms and psychiatric disorders brought limited returns
In humans, while many DNA variants can perturb the amount and biochemical function of the gene products, only a subset of them can penetrate all the intermediate levels (e.g. cells and circuits) to affect emotion and behavior. The goal of genetic studies is to understand the “architecture” of genetic influence and identify the causal variants among the many naturally occurring but phenotypically neutral variants.
Mendelian and common forms of sleep disorders cover a spectrum of genetic effects
Several Mendelian forms of sleep disorders have been described 45. A pedigree of familial advanced sleep phase disorder (FASPD) carries a S662G variant in hPer2, a human homolog of the period gene in Drosophila, a core clock gene 46. When tested in transgenic mice, the S662G change recapitulated the human phenotype 47. Similar discoveries were subsequently made for other clock genes in additional pedigrees of sleep disorders 48, 49. Meanwhile, mouse models of Clock, Npas2, Bmal1, Cry1, and Cry2 show altered sleeping traits (reviewed in 45). Collectively, these studies provided convincing support for the importance of core clock genes in regulating sleep behavior. Notably, the loss of a single clock gene in animal models is often insufficient to disrupt normal rhythms unless environmental time cues are also disrupted. In the normal environment, multiple genetic defects are often required to elicit altered behavior 10, demonstrating robustness against genetic perturbations.
In humans, the prevalence of Mendelian sleep disorders is not accurately known, but they are likely rare. The population burden of common sleep disorders is yet to be defined. Twin studies have shown that sleeping traits have a moderate genetic component in the general population, with narrow-sense heritability in the 30–40% range 50-53. Candidate gene-based association studies of common circadian rhythm disorders have focused primarily on known clock genes and have been reviewed previously 45.
Unlike candidate gene studies, genome-wide association studies (GWAS) enable an unbiased search beyond known candidates. To date, three GWAS have been conducted for sleeping traits, and have identified association signals near the NPSR1 and PDE4D genes 54, in CACNA1C, encoding an L-type calcium channel 55, and ABCC9, encoding an ATP-sensitive potassium channel 56. Attempts to replicate the CACNA1C signal have produced mixed results 55, 57. Notably, none of the core clock genes reached genome-wide significance in these GWAS, suggesting that, with the sample size currently available, the association between common variants in known clock genes and common circadian rhythm traits have not yielded replicable results.
GWAS for psychiatric disorders: Large community efforts, variable yields
An extensive literature exists regarding linkage analyses and candidate gene association studies of psychiatric traits. Taken together, candidate gene-based association for complex traits has a lackluster history, marked by poor reproducibility 58, 59. Here, I will focus on GWAS, in which the statistical threshold is higher than for candidate genes, but the signals that did achieve genome-wide significance have proven to be more reproducible. Dozens of GWAS have been conducted for five mental disorders: MDD, BD, schizophrenia, autism, and attention deficit hyperactivity disorder; and many more GWAS are added every year. A meta-analysis of MDD (11,215 patients and 9,761 controls) did not identify risk loci with genome-wide significance 60. A similar analysis of BD (11,974 patients and 51,792 controls) identified CACNA1C, a calcium channel, and ODZ4, encoding a cell surface protein 61. A meta-analysis of schizophrenia (n > 50,000 patients and controls) implicated 22 loci 62, 63. Pathway analysis showed that calcium channels are enriched in BD association intervals. Of note, the CACNA1 gene was among the top findings in the GWAS for sleeping traits described above 55, suggesting a potential common genetic link between sleep and mood disorders.
The polygenic nature of psychiatric disorders brings new challenges
A meta-analysis of GWAS for schizophrenia suggested a polygenic model, involving common alleles across thousands of loci, each contributing very small effects 64. Since then, a polygenic genetic architecture has been similarly reported for other complex traits such as multiple sclerosis and body mass index 65, 66. Combined analyses of the five psychiatric diseases mentioned above, including 33,332 cases and 27,888 controls, demonstrated strong cross-disorder genetic overlap 67, 68. Four regions, including CACNA1C, were associated with all five disorders. Among MDD, BD, and schizophrenia, risk scores calculated from polygenic signals in one disorder can explain a statistically significant (albeit small) portion of the variance of a second disorder, particularly between BD and schizophrenia, suggesting shared genetic etiology spreading numerous loci. The latest analysis of schizophrenia implicated 6,300–13,200 independent SNPs 63.
The findings described above have provided deeper insight into the genetic basis of psychiatric disorders, but they have also brought us head-on into the next puzzle: How to make use of these findings? The polygenic scores, while compelling in a statistical sense, are remarkably diffuse in a biological sense. The sheer number of risk factors has no parallel in the history of epidemiology. For a study cohort, the inherited risks are apparently spread over thousands of loci and subtly implicate numerous genes and pathways. However, for an individual patient, does he/she come to develop the disease by equally dispersed biological routes? Or, could the polygenic signal be merely illusory, formed by pooling large numbers of patients with heterogeneous causes, each of whom develop a disease due to far fewer variants, but of higher phenotypic impact 69? If the latter is true, what is the best way to identify the heterogeneous, patient-specific genetic factors? Questions such as these are the subject of ongoing debate and are discussed further in Box 1.
Box 1.
Limitations of the polygenic understanding of psychiatric disorders
Polygenicity may turn out to be the fundamental truth for most common human diseases, and its discovery may be the overriding achievement of the GWAS approach 70, 71. However, the lack of specific molecular clues remains a dark cloud over complex trait genetics, threatening its relevance as a source of useful insights. There have been two anticipated utilities of GWAS findings. The first is prediction of disease outcome – the “risk engine”, similar to an actuarial table, and the second is mechanistic understanding – the “wiring diagram” of biology 72. Currently, the predictive power of the polygenic signals is quite limited: It mostly does not outperform traditional risk factors such as family history and is unlikely to improve dramatically in the near future. Much attention has thus been turned, understandably, to the task of distilling genetic findings into pathway signals that may guide further research 70. There has been much recent progress in tool development and pathway annotation 73, 74, leading to a steady stream of pathway findings 75, 76. Pathway results, however, often lack sensitivity and specificity, because they come from (i) in silico literature mining, requiring that the connection has already been published before and is formally searchable, or (ii) enrichment analysis of generic gene annotation, which is currently incomplete, rapidly changing, and rarely addresses specifically the particular brain circuits, symptoms, or developmental stages. In this regard, new efforts to develop functional annotations for specific life stages and specific cell types represent a promising direction, and have begun to yield more disease-specific pathway findings 77.
Rare, high-impact variants take center stage
To date, genetic studies have identified ABCC9 and possibly CACNA1C for common sleeping traits, CACNA1C and ODZ4 for BD, no significant signals for MDD, 22 associated regions for schizophrenia, and broad, polygenic signals shared across multiple disorders. These GWAS, however, have not identified any core clock genes with genome-wide significance. Taken together, we currently have very few specific DNA variants with large effects to motivate experimental follow-up, especially at the interface of circadian rhythm and mood regulation. Polygenic signals and most pathway findings carry limited specificity. For example, when a psychiatric disease is said to involve hundreds of genes that, as a whole, are enriched for neurodevelopment or synaptic transmission, the notion lies closer to the question than to the answer, as it is hardly a novel insight that brain disorders will likely involve neurodevelopment and neural transmission. In the absence of a credible molecular target, these results also do not lead to clearly falsifiable predictions.
In the near future, given the paucity of unequivocal leads, it is profitable to focus on de novo, Mendelian or nearly Mendelian cases, using rare DNA variants of large impact as the source of informative genetic perturbation. The increased availability of DNA variation data in the general population, produced by large-scale DNA sequencing in studies of other human diseases, opens up opportunities of finding genetic signals of testable consequences. For example, it would be quite interesting to ask if carriers of loss-of-function mutations in core clock genes present altered sleep habits or some forms of previously unrecognized sleep disorders. If the answer is no, the next interesting question is how molecular defects in key clock genes could be compensated at the physiological level. If the answer is yes, the identified individuals, with reverse-ascertained circadian abnormalities, would provide exciting new models to probe the molecular interactions underlying circadian regulation. Logistically, this genotype-looking-for-phenotype approach will be greatly aided by a national or international registry of genetic variants of unknown functional significance. When call-back phenotyping becomes feasible, it could extend from sleep behavior to mental function parameters, providing the much needed epidemiological data to address the chicken-versus-egg question.
An integrative framework as the new frontier
In the following, I will assume that at a certain point in the future we have indeed obtained a series of DNA variants that strongly impact circadian rhythms or mood regulation, and consider the question: How can these results be useful to neurobiologists to help them explain disease mechanisms? Since we do not fully understand the components and governing principles of neural systems, a central task is to apply the right experimental tools to support a chosen level of analysis. The standard toolkit of neurobiology offers an impressive arsenal of techniques for every level. For example, electrical recording can be performed in dissociated cells, in brain slices, or in vivo. Available cell models include cultured olfactory neurons 78, skin fibroblasts 79, or induced pluripotent stem cells 80. At a higher level, brain imaging can capture morphometric or activity measures in many brain regions. The challenge at the new frontier is in integrating these tools to further a cross-level understanding: From causal genes to causal networks, with causal dynamic patterns of neural activity that are predictive of circadian rhythms and/or affective states.
How might this integration take place?
To illustrate how all the tools and data can be marshaled under an integrative “framework”, I will use a hypothetical scenario. Let us suppose that, first, genetic or gene expression data have implicated a calcium channel gene in mood disorders. And second, screening of Mendelian families or de novo cases has identified rare mutations with a strong predicted impact on channel function. Using this genetic lead, we can screen the general population to identify additional subjects carrying high-impact mutations in this gene, and in the meantime, develop targeted animal models such as gene-disrupting transgenics or optogenetic models. Using these human subjects or animal models, we can pursue the pathophysiological relevance of the mutations at multiple levels simultaneously (see Box 2). While none of the experiments in Box 2 addresses the complete mechanism, the multi-level data from the same sample series have the best chance to “fill in” the succession of gaps from genes to behavior. And the experiments, when driven by concrete genetic hypotheses, have the best chance to unravel the causal relationships in the system.
Box 2.
Simultaneous multi-level characterization of suspected high-impact genetic variants
- In a cell system, we may determine whether the risk variants alter the expression level of the channel protein or its functional properties, such as conduction capacity or ion selectivity.
- At the synaptic level, we may use brain slices to show aberrant localization of the variant protein, or demonstrate altered plasticity, e.g. stronger or weaker long-term potentiation.
- For a network-centric approach, we may find that carriers of high-risk variants exhibit altered brain activity in a specific cortical network or an abnormal connection between two such networks.
- For dynamic properties, a neural firing unit may show an atypical frequency or amplitude of oscillation, or become out of phase with another unit.
- Developmentally, there may be delayed pruning of neural processes in a specific cortical layer, or diminished neurogenesis in the adult hippocampus.
- At the behavior level, we may test sleeping traits such as the length, phase, and quality of sleep 81.
- For global gene activity profiles, we may apply transcriptomic or metabolomic analysis and identify signatures previously seen in patient samples.
- To zoom in to study a particular time period or specific cell types in vivo, we may apply optogenetic methods to control the intensity and temporal pattern of focal activation 82.
- To study genetic interaction, we may create additional transgenic rodents either in a defined background, or introduce the candidate variants into a complex background such as the high-diversity outbred lines.
Because circadian regulation represents a uniquely workable system, it is suitable as the testing ground of this integrative research program. For example, coupling between the SCN and other oscillators should be contrasted between the mutation carriers and normal controls, especially in the context of dynamic network properties. Specifically, since the master oscillator resides in a well-defined anatomical unit, the SCN, a valuable focal point of research is to interrogate the strength and phase of functional coupling between the SCN and other brain oscillators 83, 84, such as the locus coeruleus, which regulates the transition from attention to behavioral flexibility 85, or amygdala, which regulates fear, anxiety, and depression symptoms. These experiments will directly address if, and how, a single genetic mutation can affect small-circuit plasticity, temporal coordination among networks, and behavioral outcome 16.
In practice, the best efforts of cross-level integration may break down. For example, despite clear evidence for the involvement of the monoamine neurotransmitters at the cellular and pharmacological level, PET studies could not correlate depression severity to the distribution and occupancy of monoamine receptors/transporters 86. In animal models, an engineered genetic defect often fails to impinge on cellular or circuit phenotypes because of (presumably) compensatory effects of other genes. Sometimes, it takes two genetic defects to elicit a disease-like condition, but only in a certain genetic background or under certain environmental stress. In this regard, circadian regulation and its role in mood disorders present the opportunity to clarify the power and limitations of multi-scale integration.
Three hurdles: Developmental timing, nonlinear system, and ontological mismatch
The difficulty of translating genetic findings to physiological explanations is rooted in the unexplained coupling between different levels of biological organization, and the critical dimension of time, involving both developmental timing and network dynamics (Fig. 4). Neural outputs such as emotion, cognition, and behavior are not directly coded, or understood, by properties of single cells or bulk tissue averages. Rather, the brain works as spatially distributed hierarchical neural networks, molded by an individual's genetics, early development, and life experience. If much of the initial impairment takes place during a critical period of brain development, as is plausible for BD and schizophrenia, adult-onset symptoms may merely occur as aftershocks of an unobserved earlier distress. The ideal strategy for probing the temporal sequence of brain disorders is to collect the longitudinal series that covers the relevant episodes, but this is always a daunting task in human studies. In short, we still lack suitable data to leap this hurdle.
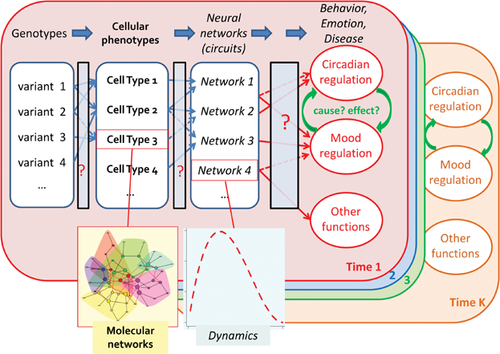
An even greater challenge lies in understanding the iterative, nonlinear interactions between genes and environment. A long-standing riddle in psychiatric illnesses is the contrast between their diverse plausible causes and the accelerated appearance of distinctive symptoms. This contrast echoes the deeper contradiction that knowledge in epidemiology is statistical, yet the practice of medicine is personal. From the statistical point of view, genetic predisposition in most cases appears to be thinly distributed over thousands of loci 63. Similarly, neural deficiencies in any population of patients implicate a wide mix of neurotransmitter systems and brain regions. However, at the individual level, the clinical presentations are clustered, distinctly recognizable, and often strongly familial, suggesting a restricted number of archetypal disturbances, possibly involving a small number of genetic origins, neurotransmitters, and brain circuits. This implies that in the natural history of a given patient, the interactions between his/her unique genes and unique environment must be dynamic, convergent, self-organizing, and self-reinforcing, bearing resemblance to the classic idea of canalization 87, 88. New analytical algorithms are needed to capture such a clearly nonlinear system, as the current case-control studies are primarily designed to screen for simple, stable effects shared across a large cohort.
Lastly, we face an ontological mismatch between the working concepts of different disciplines. As an analogy, something as simple as water has many levels of understanding: The structure of H2O does not automatically explain its taste, wetness, or how it makes the floor slippery. Likewise, the fundamental units of genetic analysis – genes, their expression patterns in time and space, and pathways – remain to be aligned with the fundamental units of neuroscience: cells, chemical synapses, local circuits, their plasticity, and the dynamics of large networks. They both remain to be connected to system-level neural correlates, such as temporal synchronization of transient neuronal assemblies. Network properties are, at least in principle, objectively measurable, yet they remain to be bridged to behavior or emotion, as subjectively acted or experienced.
Concluding remarks
The dizzying progress in the fields of genomics and neuroscience make it easy to forget that many scientific hurdles remain to be overcome. A recent reminder of these hurdles is the discussion of how psychiatric diagnosis has yet to be connected to mechanistic footing 89, 90. In this essay, I draw attention to technical limits and pressing conceptual challenges in both fields: In genetic studies, statistical evidence of association remains to be elevated to functional explanations that are experimentally testable. In neuroscience, the challenge lies in uniting the data and tools across multiple levels of analysis. In this regard, circadian regulation allows concurrent studies on multiple scales, strengthening the link between DNA variation, cellular activities, neural circuitry, its dynamics and plasticity, and mood and cognition. The analysis of the circadian-mood system provides an exceptional opportunity for developing mechanistic insights into nonlinear biological systems, and will be a testing ground for the best tools in both genomics and neuroscience. It is also suited to study the situation in which the mechanistic link breaks down. Such a case disrupts the existing best model and will help spell out how a high-level feature becomes irreducible, or how a massive amount of low-level data becomes intractable. Thus, the advances or stagnation of this research will yield valuable lessons for tackling other brain functions and other multi-level problems in biology.
Acknowledgments
I thank the past and present support from the Pritzker Neuropsychiatric Disorders Research Consortium, the Brain & Behavior Research Foundation (formerly NARSAD), and an IMHRO – Johnson & Johnson Rising Star Translational Research Award. I also thank Drs. Megan Hagenauer, Chris Gunter, and Huda Akil for providing insightful comments.
The author has declared no conflict of interest.